1. Background
Hepatitis B virus (HBV) is the most common cause of acute hepatitis and chronic liver diseases worldwide, especially in Asia and Africa (1). Almost 2 billion people around the world are infected with hepatitis B virus and 350 million of them remain chronically infected (2, 3). Studies have shown that HBV infection leads to liver damages. This situation may result in cirrhosis and hepatocellular carcinoma (HCC) in about one third of subjects with chronic HBV. The remaining two thirds are at increased risk of developing liver damage (4, 5).
Pro-inflammatory T-helper 1 (Th1) and anti-inflammatory T-helper 2 (Th2) cells regulate the cellular immune response (6). Th1 and Th2 cytokines, such as interferon gamma (IFN-γ), interleukin 6 (IL-6), transforming growth factor beta (TGF-β), interleukin 4 (IL-4) and interleukin 16 have important effect on viral hepatitis infection (7-9). In this balance, it seems that Interleukin-10 (IL-10) can play a vital role in immune response against HBV.
Interleukin-10 is produced by Th2 cells, regulatory T cells, macrophages and various cell types in the liver (including hepatocytes, Kupffer cells, sinusoidal endothelial cells and liver-associated lymphocytes). IL-10 can suppress host Th1 immune response by inhibiting the expression of pro-inflammatory cytokines such as IL-2, TNF-alpha and IFN-gamma in Th1 cells (10). IL-10 also has anti-fibrotic properties (11). In patients with chronic hepatitis, production of inappropriate amounts of IL-10 was reported to be associated with infection clearance (12, 13).
In human, the gene encoding IL-10 contains five exons and spans 5.2 kb on chromosome 1q31-q32 (14).
Recently some polymorphisms, mainly single nucleotide polymorphisms (SNPs), with biological significance have been reported in the IL-10 gene. These polymorphisms may influence the level of interleukin-10 production. Production of IL-10 is regulated at transcriptional, posttranscriptional and translational levels (15).
The polymorphisms in promoter region are more important than others, because promoter can regulate gene transcription (16-18).
Three biallelic SNPs from the transcription start site control the production of IL-10. These SNPs are located at positions –592 (rs1800872, A/C), –819 (rs1800871, T/C) and –1082 (rs1800896, A/G) (19, 20).
These SNPs form three different haplotypes of CCA, ATA and CCG. The incidence of ATG haplotype is scarce. Alleles –592C/–819C and –592A/–819T are inherited together (18, 19, 21-24). The –592C, –819C and –1082G alleles have been associated with high levels of IL-10, while CCA and ATA haplotypes produce medium and low levels of IL-10, respectively (19). Therefore, IL-10 has been implicated in the pathogenesis of HBV. It seems that allelic variation in these polymorphisms may be associated with progression of HBV infection (25). Heterogeneity in the promoter region of IL-10 gene has a role in determining the response of chronic hepatitis C to IFN-α therapy (26). Also the results suggest that IL-10 gene polymorphisms may cause an imbalance between the pro-inflammatory and anti-inflammatory cytokine responses. This phenomenon may influence susceptibility to HCV infection (27).
The natural history of HBV is different among individuals (28). Therefore, it is necessary to investigate whether IL-10 polymorphisms could serve as a suitable candidate to predict chronic HBV infection.
The association between outcomes of hepatitis B infection and these SNPs/haplotypes has been investigated in some populations (28-30). The results suggest that there is a vital role for IL-10 variations in outcomes of HBV infection. However, in some studies on polymorphism of IL-10 and HBV infection, there was no significant association (31, 32).
Turner et al. showed that the -1082 GG genotype is associated with high IL-10 production and polymorphisms at position -592 and -819 has no independent influence on IL-10 production (19).
In another study, Gao et al. indicated that IL-10 -1082 AA was associated with an increased risk of persistent HBV or HCV infection. He indicated that the IL-10 -592 AC genotype is associated with hepatocellular injury (32). Also Yan et al. showed that 592-C and 819-C alleles may increase hepatocellular injury (33).
2. Objectives
Regarding the effective role of IL10 in regulation of T-cell activity and its impact on chronic viral infections, in the present study we investigated three SNPs (rs1800872, rs1800871 and rs1800896) of the IL-10 promoter in a sample of Iranian population to determine the influence of host genetic factors in chronic HBV infection. We evaluated differences in allele, genotype and haplotype frequencies using a case-control study design.
3. Patients and Methods
3.1. Patients and Controls
To determine the association between IL-10 promoter polymorphisms and chronic HBV infection, blood samples were collected from 221 patients with chronic hepatitis B from blood transfusion organization outpatients’ clinics in Zahedan, Iran, between July and December 2014 (122 males and 99 females, aged 12 - 67 years, mean age of 29.81 ± 8.05 years old). The study was approved by the ethics committee of Zahedan University of Medical Sciences (No. 6809) and performed in Infectious diseases and tropical medicine research center, Zahedan, Iran. An informed consent was obtained from each participant.
Chronic hepatitis B was defined by positive results for HBsAg in a minimum of six months. All subjects with positive results for HBsAg with enzyme-linked immunosorbent assay (ELISA) and HBV-DNA with reverse transcription-polymerase chain reaction (RT-PCR) were selected as patients. Criteria for chronic infection of hepatitis B were as follows; HBsAg positive > 6 months, serum HBV DNA > 20,000 IU/mL and persistent or intermittent elevation in AST and ALT levels; we did not have liver biopsy results. All patients’ blood samples had an elevated alanine aminotransferase (ALT) of at least 2-fold higher than upper limits of normal value.
Exclusion criteria were coinfection with HCV, HEV, HAV or HIV, hepatorenal syndrome, hepatocellular carcinoma (HCC) and active alcohol abuse.
The control group consisted of 200 healthy subjects (109 males and 91 females, aged 17 - 67 years old, mean age of 31.11 ± 7.54 years old), none of whom had any history of hepatitis B infection. The control group comprised anti-HBs and anti-HBc positive healthy volunteers (resolved HBV). Patient and control populations were of the same geographical area. There was no difference between groups regarding gender and ethnicity.
3.2. Genotyping of Polymorphisms
Genomic DNA was extracted from a 2 mL sample of whole blood collected into EDTA. Extraction was performed using the salting-out method.
The 3 biallelic IL-10 promoter polymorphisms were detected by PCR-RFLP method using primers that amplified a short fragment of DNA containing the polymorphism.
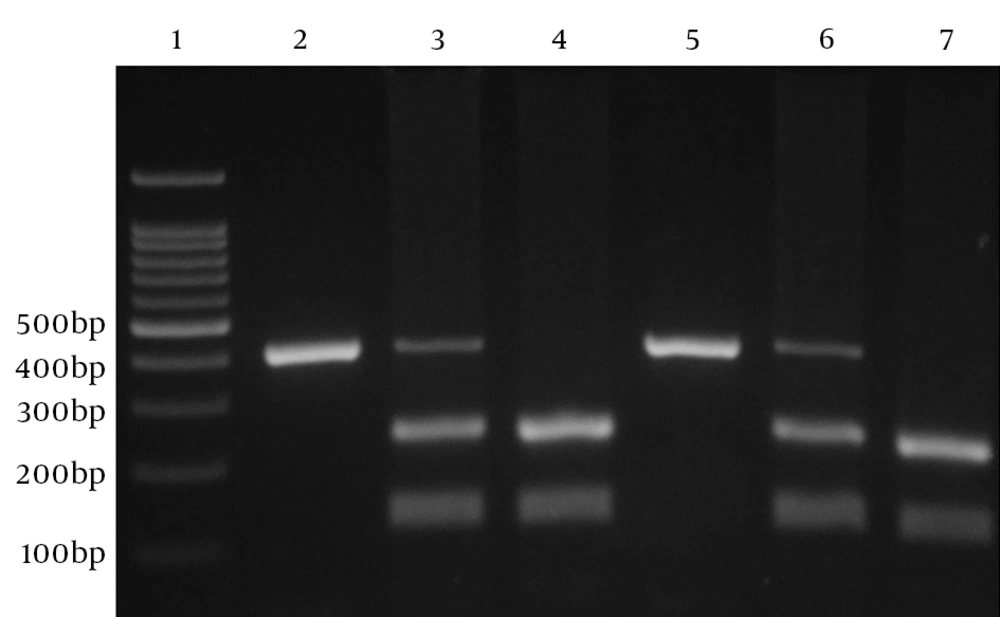
One pair of primers (forward: 5'-GGTGAGCACTACCTGACTAGC-3', reverse: 5'-CCTAGGTCACAGTGACGTGG) was used to amplify relevant fragment and analyze the 592(C/A; rs1800872) variation. Amplification was performed in a volume of 20 µL containing 1 µL of each primer, 100 ng of template DNA and 10 µL of 2X Prime Taq Premix (Genet Bio, Korea) and 7 µL ddH2O. Polymerase chain reactions were run for 30 cycles; initial denaturation for 5 minutes at 95°C, denaturation for 30 seconds at 95°C, annealing for 30 seconds at 60°C, extension for 30 seconds at 72°C and final extension for 5 minutes at 72°C. The product (10 µL) was digested with RSaI (Fermentas, Vilnius, Lithuania) restriction enzyme at 37°C for 16 hours, subjected to electrophoresis in 2% agarose gel (Invitrogen, The USA), and stained with ethidium bromide (Figure 1). IL-10 A-592 allele gives 2 fragments of 236 and 176 bp and IL-10 C-592 allele gives a single 412-bp fragment.
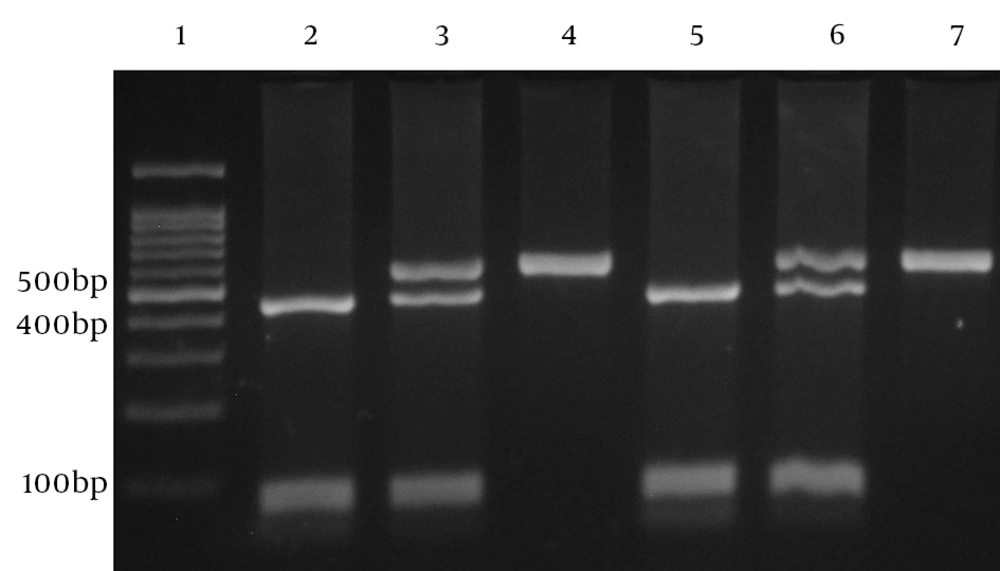
Another pair of primers (forward: 5’-CCAGATATCTGAAGAAGTCCTG-3’, reverse: 5’-TGGGGGAAGTGGGTAAGAGT) was used to amplify relevant fragment and analyze the 819 (C/T; rs1800871) variation. Amplification was performed in a volume of 20 µL containing 1 µL of each primer, 100 ng of template DNA and 10 µL of 2X Prime Taq Premix and 7 µL ddH2O. Polymerase chain reactions were run for 30 cycles: initial denaturation for 5 minutes at 95°C, denaturation for 30 seconds at 95°C, annealing for 30 seconds at 57°C, extension for 30 seconds at 72°C and final extension for 5 minutes at 72°C. The product (10 µL) was digested with RseI (Fermentas, Vilnius, Lithuania) restriction enzyme at 37°C for 16 hours, subjected to electrophoresis in 2% Agarose gel (Invitrogen, The USA) and stained with ethidium bromide (Figure 2). IL-10 C-819 allele gives 2 fragments of 443 and 116 bp and IL-10 T-819 allele gives a single 559-bp fragment.
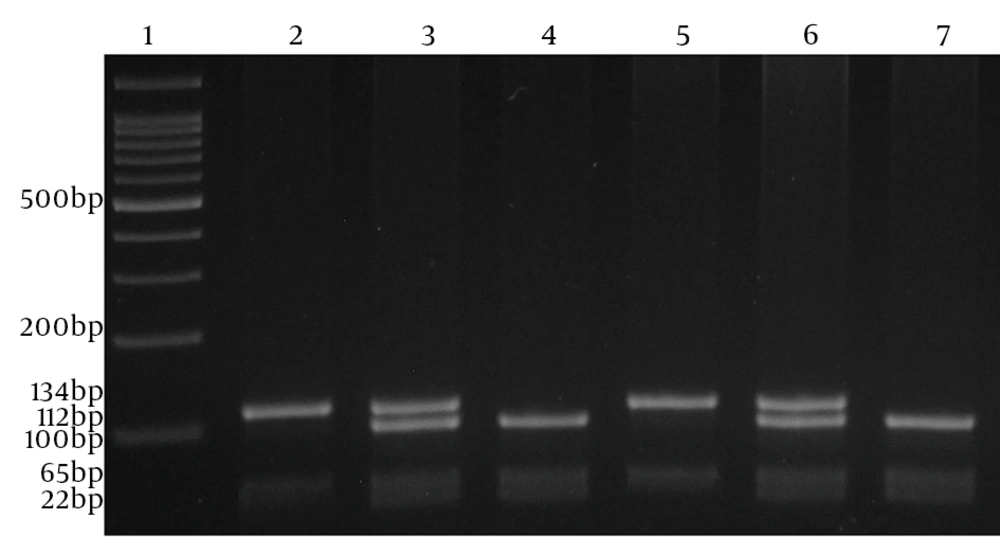
To amplify relevant fragment and analyze the 1082 (G/A; rs1800896) variation, we used this pair of primers (forward: 5’-CCAGATATCTGAAGAAGTCCTG-3’, reverse: 5’-CTCTTACCTATCCCTACTTCC). Amplification was performed in a volume of 20 µL containing 1 µL of each primer, 100 ng of template DNA, 10 µL of 2X Prime Taq Premix and 7 µL ddH2O. Polymerase chain reactions were run for 30 cycles; initial denaturation for 5 minutes at 95°C, denaturation for 30 seconds at 95°C, annealing for 30 seconds at 55°C, extension for 30 seconds at 72°C and final extension for 5 minutes at 72°C. The product (10 µL) was digested with MnlI (Fermentas, Vilnius, Lithuania) restriction enzyme at 37°C for 16 hours, subjected to electrophoresis in 4% agarose gel (Invitrogen, USA) and stained with ethidium bromide (Figure 3). IL-10 A-1082 allele gives 2 fragments of 134 and 65 bp and IL-10 G-1082 allele gives 3 fragments of 112-bp, 65 bp and 22 bp.
3.3. Statistical Analysis
Statistical analysis was performed using the statistical software package SPSS 20. The genotypic and allelic frequencies observed in patient and control groups were calculated by direct counting. The associations between allelic and genotype frequencies and HBV, as well as the odds ratio (OR) for susceptibility to infection were obtained by χ2-test and 95% confidence intervals (95% CI) from logistic regression analyses. Quantitative data presented as mean ± standard deviation. A P value of < 0.05 was considered statistically significant.
4. Results
4.1. IL-10 -592 A/C, -819 T/C, -1082 A/G Promoter Polymorphism Analysis
IL-10 SNPs -592 A/C, -819 T/C, -1082 A/G allele and genotype frequencies in HBV patients and healthy control are shown in Table 1. Genotype distribution of all polymorphisms did not deviate significantly from the Hardy-Weinberg equilibrium.
| IL10 Polymorphisms | HBV a | Control a | OR (95% CI) | P Value |
|---|---|---|---|---|
| –592 (rs1800872, A/C) | ||||
| AA | 36 (16.3) | 31 (15.5) | 1 | NA |
| AC | 168 (76.0) | 157 (78.5) | 1.08 (0.64 - 1.84) | 0.761 |
| CC | 17 (7.7) | 12 (6.0) | 0.82 (0.34 - 1.98) | 0.658 |
| AC + CC | 185 (83.7) | 169 (84.5) | 1.06 (0.63 - 1.79) | 0.825 |
| Allele | ||||
| A | 240 (54.3) | 219 (54.8) | 1 | NA |
| C | 202 (45.7) | 181 (45.2) | 1.02 (0.77 - 1.33) | 0.896 |
| –819 (rs1800871, T/C) | ||||
| TT | 40 (18.1) | 30 (15.0) | 1 | NA |
| TC | 163 (73.8) | 162 (81.0) | 1.32 (0.79 - 2.23) | 0.289 |
| CC | 18 (8.1) | 8 (4.0) | 0.59 (0.23 - 1.55) | 0.284 |
| TC + CC | 181 (81.9) | 170 (85.0) | 1.25 (0.75 - 2.10) | 0.394 |
| Allele | ||||
| T | 243 (55) | 222 (55.5) | 1 | NA |
| C | 199 (45) | 178 (44.5) | 1.02 (0.78 - 1.34) | 0.879 |
| –1082 (rs1800896, A/G) | ||||
| AA | 72 (32.6) | 100 (50.0) | 1 | NA |
| AG | 118 (53.4) | 84 (42.0) | 0.51 (0.34 - 0.77) | 0.001 |
| GG | 31 (14.0) | 16 (8.0) | 0.37 (0.19 - 0.73) | 0.004 |
| AG + GG | 149 (67.4) | 100 (50.0) | 0.48 (0.32 - 0.72) | 0.0001 |
| Allele | ||||
| A | 262 (59.3) | 284 (71.0) | 1 | NA |
| G | 180 (40.7) | 116 (29.0) | 1.68 (1.26 - 2.24) | 0.0001 |
a Values are expressed as No. (%).
-1082 G allele was significantly represented in HBV patients groups in comparison to healthy group (40.7% vs. 29%, P < 0.0001). We found higher frequencies of AG and GG genotypes at IL-10-1082 in HBV patients than healthy controls. These genotypes were associated with increased susceptibility to HBV (P = 0.001; OR = 0.51; 95% [CI], 0.34 - 0.77, P = 0.004; OR = 0.37; 95% [CI], 0.19 - 0.73).
None of the IL-10 -592 A/C and -819 T/C alleles or genotypes was associated with HBV group.
4.2. IL-10 -592 A/C, -819 T/C, -1082 A/G Haplotype Distribution
Combination of different alleles of the IL-10 SNPs -592 A/C, -819 T/C, -1082 A/G generates three haplotypes -592C,-819C,-1082G; -592C,-819C,-1082A; and -592A,-819T,-1082A (CCG, CCA, ATA respectively) (Table 2). CCG haplotype was significantly more frequent in HBV group in comparison to the control group (40.3% vs. 26.5%, P = 0.014 and appeared to be associated with susceptibility to HBV (OR = 0.56, 95% CI, 0.35 - 0.89).
a Values are expressed as No. (%).
There were no significant differences in demographic parameters between HBV cases and healthy controls (Table 3).
| Parameters | HBV, N = 221 | Control, N = 200 | P Value |
|---|---|---|---|
| Age, y a | 29.81 ± 8.05 | 31.11 ± 7.54 | 0.091 |
| Gender | 0.885 | ||
| Male | 122 | 109 | |
| Female | 99 | 91 | |
| Ethnicities | 0.817 | ||
| Sistani | 92 | 86 | |
| Baluch | 59 | 48 | |
| Others | 70 | 66 |
a Values are expressed as mean ± SD.
5. Discussion
This was the first study demonstrating the association between polymorphisms of promoter genes of IL-10 (-592 A/C, -819 T/C, -1082 A/G) and susceptibility to HBV in Iranian populations. Our strongest findings were among variants of IL-10, especially the SNP 1082 (A/G; rs1800896), which has a strong association in this sample of Iranian population. There was also an association between CCG haplotype of IL-10 and HBV infection outcome.
HBV and HCV infected individuals are living with chronic liver disease with different severity such as hepatocytes lesions, liver cirrhosis and hepatocellular carcinoma (HCC) (34).
It is believed that host genetic factors relating to genetic polymorphisms are responsible for clinical outcomes of disease (25, 28, 35). Also differences in the susceptibility to disease can be attributed to the virulence of an organism.
There was evidence that HBV infection outcome was influenced by SNP variants of the cytokines. Cytokine production differs among Individuals. This is associated with SNPs in the regulatory regions of cytokine genes (36).
IL-10 is produced by lymphocytes and monocytes and the level of its production determines immune regulation and plays an important role in hepatitis pathogenesis. Expression of IL-10 is genetically controlled. Studies have shown that IL-10 gene polymorphism is a key player in differential expression of IL-10 and would affect body’s immune response to HBV and susceptibility to HBV. Therefore, these polymorphisms can cause individually differences in immune response and lead to different host immune function (19, 37, 38). Therefore, IL-10 polymorphism determination is important for predicting susceptibility to HBV infection.
Although IL-10 variants have been associated with hepatitis C recovery (39), based on our researches, this is the first finding in Iran, since IL-10 variants have not been implicated with HBV pathogenesis before. In HCV infection some studies on various ethnic groups deny a positive association and others indicate such a link (26, 40, 41).
This study demonstrated a significant association between the IL-10 -1082 A/G polymorphism and HBV infection susceptibility in a sample of Iranian population. The other IL-10 polymorphisms were not associated with HBV infection.
In our study, only -1082 G allele was significantly represented in HBV patients. Chronic liver disease raises inflammatory and anti-inflammatory responses. These events are responsible for high IL-10 levels. Previous in-vitro studies showed that the -1082 G allele corresponds to higher IL-10 protein expression. Moreover, proinflammatory IL-10 levels increased with HBV infection progression, as compared to controls. However, the -1082 GG, AG and AA genotypes are related to high, intermediate and low IL-10 production in patients with HBV, respectively (19). The -1082 AG and GG genotypes are associated with HBV risk and can increase susceptibility to HBV in subjects. On the other hand, increased levels of IL-10 have been shown in chronic HBV patients (19), indicating that individuals carrying -1082 G allele have a risk of HBV infection. Our findings indicated that GG and AG genotypes were more prevalent in patients in comparison to healthy subjects. These results were in accordance with those reported by Turner et al., Eskdale et al. and Truelove et al. (19, 24, 42). They showed that susceptibility to HBV increases in patients with G allele and other studies in India (43) and Chinese (44) populations did not report the same result. In these populations, IL-10 -1082 A/G polymorphism was not related to chronic HBV infection.
Recently Baghi et al. (45) reported that IL10 promoter gene polymorphisms predisposed cirrhosis in infected hepatitis B patients. They showed that the -592A/C, -819T/C polymorphisms and CCG/ATA haplotype of the IL-10 gene promoter were significantly more common in patients with HBV related cirrhosis. Earlier, Sofian et al. (46) revealed that genotypes and haplotypes of IL-10 gene promoter (-1082,-819,-592) were not significantly different among controls and chronic HBV patients. They did not find any correlation between IL-10 gene promoter polymorphism and HBV infection; however, their study size was relatively small compared to our study.
These different findings are related to sample size of studied populations and/or different ethnical backgrounds. Our study was conducted in south-east of Iran. In this area, diversity of the resident population is higher than other parts of Iran and this could be the reason of these discrepancies in various studies.
However in Asian population the -1082GG genotype was rarely found (32, 47, 48). In this study, analysis of genotypes showed that subjects with -1082GG are susceptible to HBV compared to the -1082 AA genotype. In this regard, Reuses et al. noted that the -1082 GG allele produces higher levels of IL-10, which compromises immune response to infection (20).
In this study, it may indicate complexity of the immune response in humans. Also this may have been due to low number of subjects. By the way, analysis of this polymorphism in HBV infection has shown conflicting findings (28, 29, 32, 37).
It seems that IL10 variations have an important role in HBV infection. Shin et al. indicated that IL-10 -592C allele (high IL-10 producer) accelerates chronic HBV infection (49). In contrast, Cheong et al. showed that individuals with IL-10 -592A allele (low IL-10 producer) had an increased susceptibility to HBV (29). Also studies indicated that IL-10 -592AA genotype exerts a protective effect against HBV infection (19). Peng et al. showed that IL-10 intermediate producer genotypes or haplotypes cause HBeAg seroconversions in patients (28).
We did not find any significant association between IL-10 alleles, -592 A/C and -819 T/C or genotypes and susceptibility to HBV infection in patients compared to the healthy control subjects. It shows that -592 A/C and -819 T/C do not influence susceptibility to HBV infection. These two polymorphisms are in linkage disequilibrium. It means that polymorphisms at position -592 and -819 have no independent effect on IL-10 functions. It is believed that IL-10 production is independent from -592A/C and -819T/C polymorphisms (19).
In field of polymorphisms at position -592 and -819, our results were in agreement with previous studies in Chinese (32, 44) and Indian (43) populations. In Chinese researches the IL-10-592 A/C polymorphisms did not differ significantly between HBV carriers and healthy volunteers groups. However, the capacity for IL-10 production depends on genetic polymorphisms and polymorphism at position -592 of the candidate gene in patients with HBV does not emerge as a probable biomarker for determining the disease. In addition, no significant differences were detected between Indian HBV patients and healthy controls in distributions of IL-10 genotype at the −592, −819 and −1082 positions. However, according to the odds ratio it seems that heterozygosity of genotypes −592 A/C and −819 T/C is associated with liver chronicity and increased risk of persistent infection.
Conversely, Afzal et al. (27) reported that in Pakistani population, IL-10 -819 T/C and -592 A/C promoter polymorphism, at the allele level but not in genotype distribution, were significantly different in patients with hepatitis in comparison with healthy controls. In this regard we did not see any significant differences in neither alleles nor genotypes frequencies. Our results suggested lack of association of IL-10 -819 T/C and -592 A/C alleles as well as genotypes with HBV.
Wu et al. also showed that IL-10 -1082 GG genotype is associated with lower HBV viral load in comparison to patients with A allele (50). Gao et al. reported that IL-10-592 and IL-10-1082 polymorphisms are not different at the allele level between cases and control (32). A meta-analysis in 2010 indicated that IL-10-1082 polymorphism was not related to HBV infection in Asia (51). In another study conducted by Zhang et al. no significant differences were discovered in IL-10 gene promoter polymorphisms among chronic HBV patients and controls. But their study indicated that -592AA and -819TT genotypes were more prevalent in chronic HBV patients (52).
Consequently, we can obtain a suggestive evidence of association with hepatitis B for IL-10-1082 promoter polymorphism at the allele and genotype distribution, but not for IL-10-592 and -819. Some disagreements between these studies indicate the possible impact of genetic background of different populations. These studies suggest that different IL-10 gene polymorphisms can affect cytokine responses, which may in turn influence susceptibility to HBV infection.
Analysis of haplotype could explain the effect of these three SNPs on susceptibility to HBV. In previous studies, the CCG haplotype was associated with increased IL-10 production compared with the ATA and CCA haplotypes (19). In the current study, IL-10 haplotype distribution indicated that the frequency of the CCG haplotype among HBV patients was higher than that of healthy subjects.
Our findings were in agreement with the results reported by Turner (19). He indicated that the CCG haplotype frequency in hepatitis cases was higher than the controls. Nonetheless, Afzal et al. indicated significant differences between patients and controls regarding ATG and CCA haplotypes (27). Also in some studies from Caucasian, Italy and Japan, IL-10 haplotypes were not distributed differently between controls and patients (47, 49, 53). It may be due to different ethnicity background of those populations. When we analyzed haplotypes, individuals with IL-10/CCG haplotype were genetically predisposed to develop HBV. In addition, IL-10/ATA haplotype was more common in controls than patients. It means that individuals with this genetic variation are less likely to develop HBV and subjects with haplotypes having the G allele are in increased risk of HBV infection. On the other hand, several factors such as sample size, patients’ selection and different gene-gene interactions can cause conflicting results of these studies.
It seems that different cultural backgrounds such as decreasing body mass index and smoking may have effects on decreased IL-10 production (20). Moreover, epidemiological and geographical factors can cause conflicting results. Study conditions (such as number and characteristics of subjects and HBV genotype variations) can affect the results as well.
In summary, our study showed that IL-10 gene polymorphism has an influence on HBV infection. The polymorphism of IL-10 -1082 showed a significant association with susceptibility to chronic HBV. The IL-10 -1082 AG and GG were associated with increased risk of persistent HBV. IL-10 polymorphism appeared to play an essential role in human HBV susceptibility to infection. Some of our results are in agreement with earlier publications on IL-10 promoter polymorphism and HBV infection. Other observations from this study are contrary to previous findings in other populations. There are different IL-10 binding receptors and homologues to IL-10 (54). So it is difficult to specify the real effect of genetic polymorphisms on disease susceptibility. These phenomena can affect the association between IL-10 promoter polymorphisms and HBV infection. Further researches on functional implications of IL10, specifically regarding immune response to HBV, are warranted. Moreover, we believe that our findings may stimulate further investigations on a larger size to assess the association of these polymorphisms in HBV patients.