1. Background
Hepatitis C Virus (HCV) has infected about 170 million patients worldwide and it has been considered as a major cause of liver failure and Hepatocellular Carcinoma (HCC) (1). Patients with transfusion-dependent thalassemia are at higher risk of acquiring HCV infection, especially those who received their first transfusion in Iran before the introduction of the HCV donor - screening program in 1995 (2). Hepatitis C virus seroprevalence in Iranian thalassemic patients is approximately 18% (3). Liver failure as the consequence of HCV infection is the second cause of mortality in thalassemic patients (2). Iron overload following frequent blood transfusion deteriorates liver fibrosis in these patients (4). Antiviral therapy and achievement of Sustained Virological Response (SVR) can act to avoid liver failure and HCC, and can also decrease reservoir of HCV infection in thalassemic patients. Combination of Pegylated-Interferon (PEG-IFN) and Ribavirin (RBV) has been established as the standard anti - HCV therapy. Although RBV is not approved by the food and drug administration (FDA) for patients with thalassemia because of increasing hemolytic anemia, there are some published data regarding the safety and increment of SVR following administration of RBV in patients with thalassemia (5, 6). However, patients with thalassemia have some comorbidities due to iron overload and PEG-IFN/RBV combination therapy. Moreover, treatment with PEG-IFN and RBV in patients with thalassemia is associated with more adverse effects and more needs of transfusion (7). On the other hand, the mono - therapy with PEG-IFN resulted in low SVR rate in thalassemic patients with HCV genotype-1 infection, which seems to be caused by liver iron overload and absence of RBV (5).
Numerous factors including gender, age, HCV RNA level prior to treatment, stage of liver fibrosis, and HCV genotypes have been shown to determine the success rate to antiviral therapy (8). In addition, a number of recently published Genome-Wide association studies (GWAS) have shown that polymorphisms within the interferon lambda (IFNL) genomic region result in differences between patients’ responses to anti - HCV treatment (9-11). Among these single nucleotide polymorphisms (SNPs), rs12979860 (located 3 kb upstream of the IL28B (IFNL3) gene in intron 1 of IFNL4 gene) and rs8099917 (located 8 kb upstream of the IFNL3 gene) have been identified as two predictive factors of response to antiviral therapy. Given that antiviral therapy in thalassemic patients with hepatitis C is more difficult than in non-thalassemic patients, detection of predictive factors for SVR is necessary. Unfortunately, there are currently no reports in the literature regarding the role of these SNPs in thalassemic patients with HCV infection, who were treated with PEG-IFN and RBV.
2. Objectives
We aimed to assess the impact of rs12979860 and rs8099917 polymorphisms on the rate of response to combination therapy with PEG-IFN and RBV in thalassemic patients with chronic hepatitis C (CHC).
3. Materials and Methods
3.1. Study Population
In this cross - sectional study from 2010 to 2013, 143 transfusion-dependent thalassemic patients, who had HCV RNA > 50 IU/mL for more than six months were evaluated and treated at the Tehran Hepatitis Clinic a clinic associated with the Baqiyatallah Research Center for Gastroenterology and Liver Diseases (BRCGL). Patients who were co-infected with hepatitis B virus and human immunodeficiency virus, decompensated cirrhotic patients, cases with HCC, and those who had contraindication for interferon therapy including severe heart failure, poorly controlled diabetes mellitus, poorly controlled psychiatric disorder, and also, patients with baseline hemoglobin below 8 g/dL were excluded from the study. All patients had no history of previous treatment with PEG-IFN/RBV. All study participants provided an informed consent and the study design was approved by the ethics committee of BRCGL. The study protocol conforms to the ethical guidelines of the 1975 declaration of Helsinki.
3.2. Treatment Regimen and Response Definition
All of the patients were treated with combination of PEG-IFN-α-2a (Pegasys, Roche, Basel, Switzerland) and RBV (Copegus, Roche, Basel, Switzerland) or PEG-IFN-α-2b (Pegintron, Schering - plough, Puerto Rico, USA) and RBV (Rebetol, Schering - plough, Puerto Rico, USA). The dose of Pegasys was 180 μg subcutaneously once a week in combination with oral RBV 600 - 1000 mg per day according to patients’ weight and hemoglobin level. The dose of Pegintron was 80, 100 or 120 μg subcutaneously once a week according to patients’ weight in combination with oral RBV 600 - 1000 mg per day according to patients’ weight and hemoglobin level. The treatment duration was 24 weeks for HCV genotype-3 infection or 48 weeks for HCV genotype-1 infection, which could be extended regarding patient’s response and compliance. In patients with HCV genotype-1 infection, who achieved partial early virological response, the treatment course was extended to 72 weeks. All patients who were infected with mixed HCV genotypes had HCV genotype-1 as a component of the HCV pool and as a result they were treated the same way as patients with HCV genotype-1 infection and were considered as HCV genotype-1- infected patients in the subsequent analysis.
Serum HCV RNA level was measured at weeks 4, 12, 24, 48 and 72 during treatment and 24 weeks after treatment cessation. Undetectable HCV RNA (< 10 IU/mL) at the end of four weeks of treatment was considered as Rapid Virological Response (RVR). Early Virological Response (EVR) was defined as undetectable HCV RNA at the end of week 12 (complete EVR, cEVR) or at least 2 - log decrease in HCV RNA at the end of week 12 in comparison with the baseline HCV RNA level (partial EVR, pEVR). Undetectable HCV RNA six months after the treatment cessation was considered as SVR, which determined the treatment success. Treatment non-response was defined as < 2 -log decrease in HCV RNA level at week 12, compared to the baseline or detectable HCV RNA at week 24. If undetectable level of HCV RNA was achieved at the end of treatment and the HCV RNA became detectable six months later, the patient was considered as a relapse case.
3.3. Laboratory and Liver Fibrosis Assessments
Assessment of HCV RNA level was carried out using COBAS® TaqMan® HCV Test v2.0 (Roche Diagnostics), according to the manufacturer’s instructions. In this study, rs12979860 and rs8099917 SNPs were assessed as the most common IFNL polymorphisms. The detailed protocol of the polymerase chain reaction - restriction fragment length polymorphism (PCR-RFLP) method for genotyping of rs12979860 and rs8099917 SNPs has been previously described (12). Briefly, genomic DNA was extracted from patients’ peripheral blood using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The PCR was performed using Accupower PCR PreMix (Bioneer Corp., Daejeon, South Korea) with the following conditions: 94°C for five minutes, 35 cycles of 94°C for 20 seconds, 66°C for 20 seconds, and 72°C for 20 seconds, followed by 72°C for five minutes. Primers used for the reaction included: rs12979860, 5’ - GCGGAAGGAGCAGTTGCGCT - 3’ and 5’-GGGGCTTTGCTGGGGGAGTG-3’; or rs8099917, 5’-CCCACTTCTGGAACAAATCGTCCC-3’ and 5’-TCTCCTCCCCAAGTCAGGCAACC-3’. Next, the PCR amplicons were digested for more than one hour with 10 U of restriction endonuclease: BstUI for rs12979860 or BsrDI for rs8099917 (Fermentas of Thermo Fisher Scientific, Waltham, MA, United States). The digested PCR products were separated on 3% agarose gels revealing the following sized fragments: rs12979860, 196 and 45 bp for the CC genotype, 241, 196 and 45 bp for the CT genotype, or 241 bp for the TT genotype; rs8099917, 552 bp for the TT genotype, 552, 322 and 230 bp for the GT genotype, and 322 and 230 bp for the GG genotype. Liver specimens were processed using standard criteria. Grading of inflammation and staging of fibrosis were determined by the modified Knodell scoring system (13). Grade on liver biopsy of ≤ 6 was considered as mild, 7 - 12 as moderate, and ≥ 13 as severe liver necro-inflammation. Stage on liver biopsy of ≤ 2 was considered as mild, 3 - 4 as moderate, and ≥ 5 as severe liver fibrosis (cirrhosis). Liver histology was evaluated by an expert pathologist, who was blind to the study design.
3.4. Statistical Analysis
Categorical variables were expressed by frequencies and percentages. Continuous variables with normal distribution were expressed by mean ± Standard Deviation (SD) and continuous variables deviated from the normal distribution by median (interquartile range). Fisher exact test was used for analysis of categorical variables, t - test for continuous variables with normal distribution, and Mann - Whitney U test for continuous variables deviated from normal distribution. The Hardy - Weinberg Equilibrium (HWE) was assessed for both rs12979860 and rs8099917 SNPs, and the Linkage Disequilibrium (LD) between these SNPs was calculated. All baseline variables with a P < 0.1 in univariate analysis were entered to a logistic regression model. To prevent multi-collinearity resulted by the LD between the two SNPs different logistic regression models with inclusion of a single SNP were considered (14). P values of less than 0.05 were considered to be statistically significant. Statistical analysis was performed using the SPSS software version 20. Statistical graphs were generated using the GraphPad Prism software version 6.
4. Results
4.1. Patients’ Baseline Characteristics
The patients’ characteristics are summarized in Table 1. In this study, 143 thalassemic patients with mean age of 24.47 were included; most of them were infected with HCV genotype-1 (61.5%) followed by HCV genotype-3 (35.7%). The most frequent rs12979860 genotype was CT (50.3%) and the rs8099917 genotype was TT (60.7%) (Table 1). The distribution of both rs12979860 and rs8099917 genotypes were in HWE (P = 0.50 and P = 0.36, respectively). On the other hand, the two SNPs were in a moderate LD (D’ = 1.0, r2 = 0.44).
| Variables | All Patients (n = 143) a |
|---|---|
| Gender | |
| Male | 86 (60.1) |
| Female | 57 (39.9) |
| Age, y | 24.47 ± 5.60 |
| Range | |
| Min | 10 |
| Max | 43 |
| BMI b | |
| Median, IQR | 19.98 (4.1) |
| Range | |
| Min | 14.4 |
| Max | 26.8 |
| Serum ALT, IU/L b | |
| Median, IQR | 64 (62) |
| Range | |
| Min | 6 |
| Max | 272 |
| Serum AST, IU/L b | |
| Median, IQR | 59 (53) |
| Range | |
| Min | 6 |
| Max | 371 |
| Liver fibrosis, stage b | |
| Mild | 35 (25.9) |
| Moderate | 45 (33.4) |
| Severe | 55 (40.7) |
| Liver inflammation, grade b | |
| Mild | 72 (53.3) |
| Moderate | 51 (37.8) |
| Severe | 12 (8.9) |
| HCV RNA, Log IU/mL b | |
| Median, IQR | 5.75 (5.95) |
| Range | |
| Min | 3.21 |
| Max | 7.22 |
| HCV genotype | |
| 1a | 84 (58.7) |
| 1b | 4 (2.8) |
| 3a | 51 (35.7) |
| Mixed genotypes | 4 (2.8) |
| rs12979860 | |
| CC | 51 (35.7) |
| CT | 72 (50.3) |
| TT | 20 (14.0) |
| rs8099917 b | |
| TT | 82 (60.7) |
| GT | 44 (32.6) |
| GG | 9 (6.7) |
aValues are expressed as No. (%) or mean ± SD.
bThe data were missed in less than 10% of patients.
4.2. Hepatitis C Treatment Response in Patients With Thalassemia
In the present study, 95 (66.4%) patients received PEG-IFN-α-2a and 48 (33.6%) were treated with PEG-IFN-α-2b. Treatment was prolonged for 72 weeks in 11 (12.0%) patients with HCV genotype-1 infection regarding pEVR achievement. Treatment was withdrawn in 25 (27.2%) patients with HCV genotype-1 infection and in six (11.8%) patients with HCV genotype-3 infection because of breakthrough or non-response. Treatment course was not withdrawn in any patients due to adverse events or non-compliance.
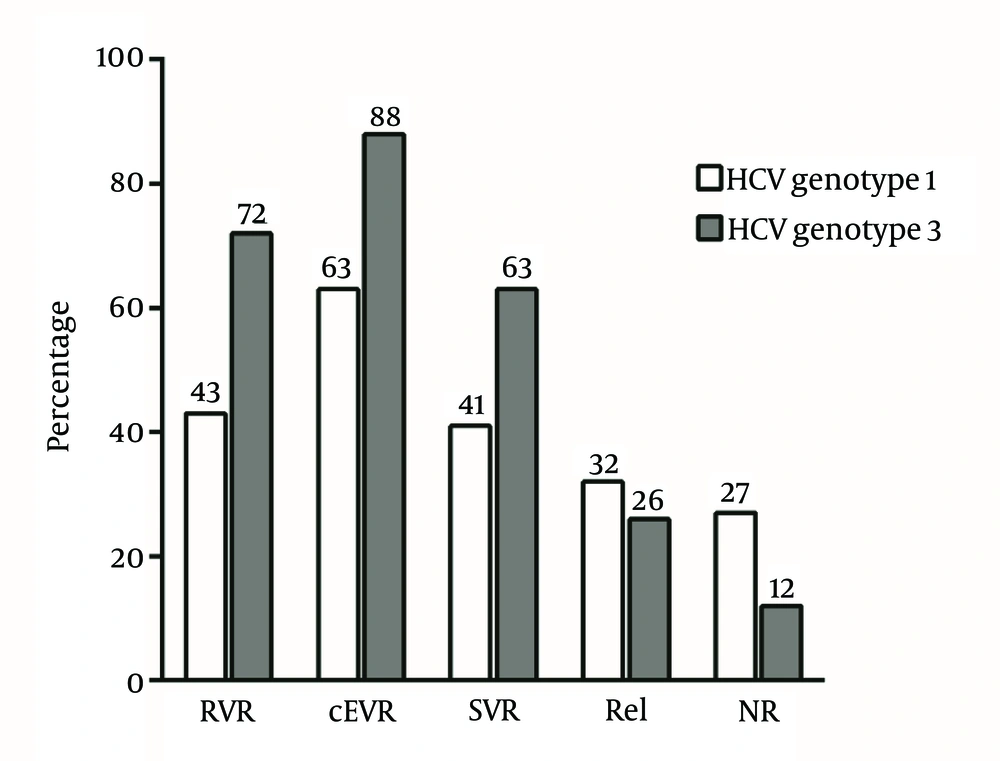
Within the cohort of 143 patients, 70 (49.0%) had SVR, 31 (21.7%) non-response or breakthrough and 42 (29.4%) relapse. The rate of SVR was higher in patients, who were treated with PEG-IFN-α-2b compared to the response rate of the patients, who received PEG-IFN-α-2a; however, the difference was not statistically significant. Among 11 patients with pEVR, who underwent 72 weeks of prolonged treatment, three (27.3%) achieved SVR. The rates of RVR, cEVR, SVR, relapse and non-response in relation to HCV genotypes are shown in Figure 1. The rate of virological responses was significantly lower in patients with HCV genotype-1 infection than that of patients with HCV genotype-3 infection, including RVR (P = 0.015; OR = 0.3; CI = 0.12 - 0.75), cEVR (P = 0.001; OR = 0.22; CI = 0.09 - 0.58) and SVR (P = 0.015; OR = 0.42; CI = 0.21 - 0.84). Furthermore, the rate of non-response was significantly higher in patients with HCV genotype-1 infection than in patients with HCV genotype-3 infection (P = 0.035; OR = 2.78; CI = 1.33 - 8.33) yet the relapse rate was not significantly different among patients with HCV genotype-1 and HCV genotype-3 infection (P = 0.57; OR = 1.35; CI = 0.63 - 2.86).
4.3. Factors Associated With Treatment Response in Thalassemic Patients With Hepatitis C Virus Genotype-1 Infection
Among baseline parameters, HCV RNA level of < 600 000 IU/mL, rs12979860 CC and rs8099917 TT genotypes were significantly correlated with achievement of cEVR in patients with HCV genotype-1 infection (Table 2). Multivariate analysis of these parameters showed that rs12979860 and rs8099917 SNPs and HCV RNA level remained as predictors of cEVR in thalassemic patients with HCV genotype-1 infection (Table 3). Furthermore, among baseline predictors, rs12979860 and rs8099917 SNPs were found to be the only parameters associated with achievement of SVR in thalassemic patients with HCV genotype-1 infection (Table 4). On the other hand, the on - treatment responses including RVR and cEVR were significantly associated with achievement of SVR in HCV genotype-1 infection; odds ratio (OR) of 4.89 and 16.44, respectively (Table 4). The sensitivity, specificity, Positive Predictive Value (PPV) and Negative Predictive Value (NPV) of rs12979860 CC genotype for prediction of SVR were 55.3%, 79.6%, 65.6% and 71.7%, respectively.
| Variables | cEVR (n = 57) | Non - cEVR (n = 34) | OR (95% CI) | P Valuea |
|---|---|---|---|---|
| Gender | 0.825 | |||
| Female | 22 (38.6) | 12 (35.3) | Ref. | |
| Male | 35 (61.4) | 22 (64.7) | 0.87 (0.36 - 2.10) | |
| Age, y | 0.665 | |||
| > 24 | 28 (49.1) | 19 (55.9) | Ref. | |
| < 24 | 29 (50.9) | 15 (44.1) | 1.31 (0.56 - 3.08) | |
| Cirrhosis b | 0.074 | |||
| Yes | 18 (34.0) | 18 (54.5) | Ref. | |
| No | 35 (66.0) | 15 (45.5) | 2.33 (0.96 - 5.68) | |
| Liver necro - inflammation grade b | 0.180 | |||
| Moderate + Severe | 20 (37.7) | 18 (54.5) | Ref. | |
| Mild | 33 (62.3) | 15 (45.5) | 1.98 (0.82 - 4.78) | |
| HCV RNA level b | 0.026 | |||
| > 600000 | 24 (42.1) | 22 (68.8) | Ref. | |
| < 600000 | 33 (57.9) | 10 (31.3) | 3.03 (1.21 - 7.54) | |
| rs12979860 | 0.002 | |||
| CT + TT | 30 (52.6) | 29 (85.3) | Ref. | |
| CC | 27 (47.4) | 5 (14.7) | 5.22 (1.77 - 15.41) | |
| rs8099917 b | 0.003 | |||
| GT + GG | 16 (28.6) | 19 (63.3) | Ref. | |
| TT | 40 (71.4) | 11 (36.7) | 4.32 (1.68 - 11.08) |
aFisher - exact test.
bThe data were missed in less than 10% of patients.
| Variables | Logistic Regression Model 1 | Logistic Regression Model 2 | ||
|---|---|---|---|---|
| Adjusted OR (95% CI) | P Value | Adjusted OR (95% CI) | P Value | |
| Absence of cirrhosis | 2.01 (0.76 - 5.31) | 0.161 | 1.90 (0.68 - 5.34) | 0.222 |
| HCV RNA level < 600000, IU/mL | 2.78 (1.03 - 7.53) | 0.044 | 3.40 (1.15 - 10.07) | 0.027 |
| rs12979860 CC genotype | 5.27 (1.67 - 16.63) | 0.005 | NA | NA |
| rs8099917 TT genotype | NA | NA | 4.44 (1.56 - 12.69) | 0.005 |
| Variables | SVR (n = 38) | Non - SVR (n = 54) | OR (95%CI) | P Valuea |
|---|---|---|---|---|
| Gender | 0.124 | |||
| Female | 18 (47.4) | 16 (29.6) | Ref. | |
| Male | 20 (52.6) | 38 (70.4) | 0.47 (0.20 - 1.11) | |
| Age, y | 0.672 | |||
| > 24 | 18 (47.4) | 29 (53.7) | Ref. | |
| < 24 | 20 (52.6) | 25 (46.3) | 1.29 (0.56 - 2.96) | |
| Cirrhosis b | 0.270 | |||
| Yes | 12 (33.3) | 24 (47.1) | Ref. | |
| No | 24 (66.7) | 27 (52.9) | 1.78 (0.73 - 4.31) | |
| Liver necro - inflammation grade b | > 0.999 | |||
| Moderate + Severe | 16 (44.4) | 23 (45.1) | Ref. | |
| Mild | 20 (55.6) | 28 (54.9) | 1.02 (0.43 - 2.42) | |
| HCV RNA level b | 0.289 | |||
| > 600000 | 17 (44.7) | 29 (56.9) | Ref. | |
| < 600000 | 21 (55.3) | 22 (43.1) | 1.63 (0.70 - 3.79) | |
| rs12979860 | 0.001 | |||
| CT + TT | 17 (44.7) | 43 (79.6) | Ref. | |
| CC | 21 (55.3) | 11 (20.4) | 4.83 (1.92 - 12.12) | |
| rs8099917 b | 0.046 | |||
| GT + GG | 10 (27.0) | 25 (50.0) | Ref. | |
| TT | 27 (73.0) | 25 (50.0) | 2.70 (1.08 - 6.73) | |
| RVR c | 0.007 | |||
| No | 10 (37.0) | 23 (74.2) | Ref. | |
| Yes | 17 (63.0) | 8 (25.8) | 4.89 (1.59 - 14.99) | |
| cEVR b | < 0.001 | |||
| No | 3 (7.9) | 31 (58.5) | Ref. | |
| Yes | 35 (92.1) | 22 (41.5) | 16.44 (4.48 - 60.29) |
aFisher - exact test.
bThe data were missed in less than 10% of patients.
cThe data were missed in more than 10% of patients.
4.4. Factors Associated With Treatment Response in Thalassemic Patients With Hepatitis C Virus Genotype-3 Infection
In the group with HCV genotype-3 infection, the number of patients who did not achieve cEVR was very small (six cases) and as a result the statistical power was too low to assess the parameters influencing achievement of cEVR. Among baseline parameters, female sex and age of < 24 were associated with achievement of SVR in patients with HCV genotype-3 infection (Table 5), however in multivariate analysis, only age of < 24 remained as a predictor of SVR in these patients (Table 6). Moreover, RVR and cEVR were both associated with achievement of SVR with OR of 16.62 and 11.07, respectively (Table 5).
| Variables | SVR (n = 32) | Non - SVR (n = 19) | OR (95 %CI) | P Valuea |
|---|---|---|---|---|
| Gender | 0.047 | |||
| Female | 18 (56.3) | 5 (26.3) | Ref. | |
| Male | 14 (43.7) | 14 (73.7) | 0.28 (0.08 - 0.96) | |
| Age, y | 0.020 | |||
| > 24 | 14 (43.8) | 15 (78.9) | Ref. | |
| < 24 | 18 (56.2) | 4 (21.1) | 4.82 (1.31 - 17.79) | |
| Cirrhosis b | 0.762 | |||
| Yes | 11 (36.7) | 8 (44.4) | Ref. | |
| No | 19 (63.3) | 10 (55.6) | 1.38 (0.42 - 4.54) | |
| Liver necro - inflammation grade b | 0.766 | |||
| Moderate + Severe | 14 (46.7) | 10 (55.6) | Ref. | |
| Mild | 16 (53.3) | 8 (44.4) | 1.43 (0.44 - 4.62) | |
| HCV RNA level | 0.145 | |||
| > 600000 | 11 (34.4) | 11 (57.9) | Ref. | |
| < 600000 | 21 (65.6) | 8 (42.1) | 2.63 (0.82 - 8.43) | |
| rs12979860 | 0.247 | |||
| CT + TT | 18 (56.3) | 14 (73.7) | Ref. | |
| CC | 14 (43.7) | 5 (26.3) | 2.18 (0.63 - 7.50) | |
| rs8099917 b | > 0.999 | |||
| GT + GG | 11 (37.9) | 7 (36.8) | Ref. | |
| TT | 18 (62.1) | 12 (63.2) | 0.96 (0.29 - 3.16) | |
| RVR c | 0.003 | |||
| No | 2 (9.5) | 7 (63.6) | Ref. | |
| Yes | 19 (90.5) | 4 (36.4) | 16.62 (2.47 - 111.8) | |
| cEVR | 0.022 | |||
| No | 1 (3.1) | 5 (26.3) | Ref. | |
| Yes | 31 (96.9) | 14 (73.7) | 11.07 (1.81 - 103.78) |
aFisher - exact test.
bThe data were missed in less than 10% of patients.
cThe data were missed in more than 10% of patients.
| Variables | Adjusted OR (95%CI) | P Value |
|---|---|---|
| Age < 24, y | 4.92 (1.26 - 19.19) | 0.022 |
| Female gender | 3.70 (0.99 - 13.70) | 0.051 |
4.5. Sustained Virological Response, Virological Non-response, Relapse and rs12979860 in Thalassemic Patients With Hepatitis C Virus Infection
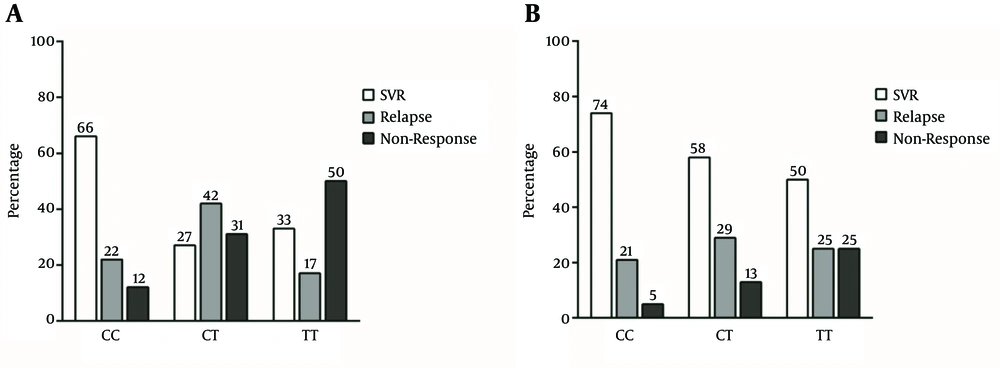
In HCV genotype-1 infection, the majority of patients with rs12979860 CC genotype achieved SVR while the majority of patients with rs12979860 CT genotype relapsed and the majority of patients with rs12979860 TT genotype were non-responders (P linear - by - linear association = 0.002) (Figure 2A). In HCV genotype-3 infection, the highest SVR rate was observed in rs12979860 CC genotype and there was a trend in the decrease of SVR in CT and TT genotypes. Furthermore, in the group with HCV genotype-3 infection, the lowest rate of non-response was observed in patients with rs12979860 CC genotype, which gradually increased in CT and TT genotypes (P linear-by-linear association = 0.122) (Figure 2B).
5. Discussion
In the present study, the prevalence of rs12979860 and rs8099917 genotypes in patients with thalassemia was similar to that of the other Iranian CHC patients. Given that Iran is located in the Middle East region with Caucasian ethnicity and based on the results of previous studies the major rs12979860 genotype was CT followed by CC and TT, and also the major rs8099917 genotype was TT followed by GT and GG in patients with CHC (15-17). The most prevalent HCV genotype in the patients with thalassemia of the present study was HCV genotype-1a followed by HCV genotype-3a that was similar to other studies on patients with and without thalassemia in Iran (18-20).
In this study, the response rate to PEG-IFN and RBV was 41.3% in thalassemic patients with HCV genotype-1 infection and 62.8% in thalassemic patients with HCV genotype-3 infection. These data were similar to the results of a recent meta - analysis, which showed that SVR rate in thalassemic patients with HCV genotype-1, who were treated with conventional-or PEG-IFN and RBV, was lower than its rate in thalassemic patients with HCV genotype-3 (21). A study on 280 thalassemic patients with CHC, who were treated with PEG-IFN monotherapy or PEG-IFN and RBV, showed that the addition of RBV to PEG-IFN increased the rate of SVR especially in patients with HCV genotype-1 (5). However, the rate of RBV-associated anemia and the need for transfusion were more in patients with thalassemia who were treated with combination of PEG-IFN and RBV versus patients with thalassemia who were treated with PEG-IFN monotherapy (5). Therefore, it was recommended to add low - dose RBV to PEG-IFN therapy in thalassemic patients considering their clinical situation. Moreover, it seems that the rate of SVR in patients with thalassemia, who were treated with combination of PEG-IFN and RBV, was lower than non-thalassemic patients due to suboptimal dose of RBV and iron overload in the liver (5, 22, 23). Furthermore, the rate of treatment side - effects, RBV dose reduction and discontinuation of therapy due to side-effects in thalassemic patients with CHC are often more than non-thalassemic patients. Overall, antiviral therapy in transfusion-dependent thalassemic patients with HCV infection is conflicting. In the present study, 40.7% of patients had severe fibrosis at the time of therapy. Iron overload in thalassemic patients with CHC aggregates liver fibrosis and many of these patients have cirrhosis at the time of therapy, so the risk of liver decompensation should be considered in these patients during antiviral therapy. In addition, other adverse effects of iron overload such as heart failure, diabetes mellitus and hypothyroidism were observed more frequently in patients with thalassemia, which can cause more difficulties during PEG-IFN therapy (24). On the other hand, monotherapy with PEG-IFN has low response rate, and the combination of PEG-IFN and RBV therapy significantly results in higher rate of SVR, however we should note that the addition of RBV can increase blood transfusion frequencies and thus may aggregate the adverse effects of iron overload especially heart failure. Regarding these facts, the threshold of starting HCV antiviral therapy with PEG-IFN and RBV is different in patients with thalassemia compared to other patients and also its adverse effects are more life threatening. Even in some patients with major co-morbidities, the physician should compare the risk of antiviral therapy to the risk of liver failure before starting therapy. As a result, the detection of predictive factors of SVR is very important in patients with thalassemia.
Based on the previous studies on non-thalassemic patients, who were treated with PEG-IFN and RBV, several factors such as female gender, younger age, low stage of liver fibrosis, low level of pretreatment HCV RNA, HCV genotypes-2 and-3 and host genetics such rs12979860 CC and rs8099917 TT genotypes have been known as the favorable predictive factors of SVR (25-27). However, the results of this study showed that rs12979860 CC and rs8099917 TT genotypes were the only predictive factors of SVR in patients with thalassemia, who were infected with HCV genotype-1. In contrast to non-thalassemic patients, there was no association between pretreatment HCV RNA level and stage of liver fibrosis with SVR in these patients. Moreover, two previous studies showed that pretreatment HCV RNA level and stage of fibrosis were not associated with SVR in patients with thalassemia (5, 28). Iron overload in patients with thalassemia may explain this finding. Similar to previous studies, the effect of on - treatment response (RVR and cEVR) was more important than the baseline predictors.
There are no studies in the literature that have assessed the association between polymorphisms near the IL28B gene and SVR in patients with thalassemia, who were treated with PEG-IFN and RBV. A study on patients with thalassemia, who were treated with conventional-IFN monotherapy, showed that the TT genotype of rs8099917 and CC genotype of rs12979860 polymorphisms were associated with SVR (28). Moreover, surprising data showed that NPV of rs12979860 for SVR achievement was 71.7%, which means that the majority of patients with non - CC genotypes of rs12979860 did not achieve SVR. However, high NPV of rs12979860 SNP in patients with thalassemia may motivate physicians to treat patients with rs12979860 non - CC genotypes with new Direct-acting Antiviral (DAA) agents with fewer side effects.
There was no relationship between polymorphisms near the IL28B gene and the response rate in thalassemic patients with HCV genotype-3 infection. This may be due to the small sample size and more studies should be conducted. A recent meta-analysis on non-thalassemic patients with hepatitis C genotype-3 infection showed that the TT genotype of rs8099917 and CC genotype of rs12979860 were the predictors of SVR and RVR in Caucasian patients, who were treated with combination of PEG-IFN and RBV (29).
The first generation of DAAs, including NS3/4A Protease Inhibitors (PIs) (Telaprevir or Boceprevir), have been proved effective in CHC patients, who were infected with HCV genotype-1 in 2011. However, these medications cause more severe anemia than dual therapy with PEG-IFN and RBV, so these drugs cannot be used for patients with thalassemia. Fortunately, several new DAAs, including second - wave and second - generation NS3, NS5A and NS5B inhibitors are approved for the treatment of HCV infection. They have higher antiviral potency, fewer adverse effects and shorter treatment duration compared to Telaprevir and Boceprevir regimens and some of them have pan - genotypic effects while their efficacy is not related to the host genetics (30). Furthermore, newer regimens are interferon - and ribavirin - free (31, 32). In recent findings, these treatments did not result in hemolysis and anemia as an adverse side effect and also these medications were used in cirrhotic patients. However, these new DAAs regimens have several and considerable drug - drug reactions (33). We should note that the majority of thalassemic patients with hepatitis C live in developing countries and these newer regimens might not be affordable soon. In addition, these new treatments were not evaluated in trials for patients with thalassemia and their safety is unknown for these patients. It should be considered that many of these patients have cardiac disease secondary to iron overload and they use cardiac drugs, however their drug reactions with new DAAs is unknown.
In conclusion, polymorphisms near the IL28B gene were the only predictive factor of response to treatment with PEG-IFN and RBV regimen in patients with thalassemia and as a result we suggest the identification of these polymorphisms in all of thalassemic patients with CHC genotype-1 before making decisions on antiviral therapy. In thalassemic patients with rs12979860 CC genotype and without severe comorbidities, the physician can motivate them to start combination therapy with PEG-IFN and RBV regimen as soon as possible. Upon availability of newer DAAs with high cost in developing countries, it is rational to try the dual therapy before using new DAAs in these patients. Based on high virological response rate to PEG-IFN and RBV combination therapy in patients infected with HCV genotype-3, we can treat these patients with PEG-IFN and RBV combination therapy, regardless of IFNL polymorphisms.