1. Background
Hepatitis C virus (HCV) is a global healthcare problem, with a prevalence of around 3% worldwide (1); it has six major genotypes (types 1 – 6). Most infected patients will establish chronicity with long-term complications, including liver fibrosis, cirrhosis, and hepatocellular carcinoma (HCC) (2). The success rates of current treatments with interferon (IFN)-based therapy are highly variable and depend on both host and viral factors (3).
Compared to other developing countries, Egypt has a high prevalence of HCV, where 20% of Egyptian blood donors are seropositive for HCV antibodies (1). Approximately 90% of Egyptian HCV patients belong to genotype-4. Although HCV-4 causes about 20% of chronic hepatitis C in the world, it has not been subjected to enough research, most probably due to its restricted localization to the Middle East and Africa, where the management strategies for infected patients are not as well developed as for genotype-1, -2, and -3 (4, 5).
MicroRNAs are endogenous small non-coding RNAs (≈ 22 nucleotides) involved in the regulation of many cellular processes (6). Several host miRNAs have been suggested to be involved in HCV entry, the establishment of viral infection, and the multi-step process of chronic HCV infection. The ability of the virus to take over certain cellular miRNAs and its persistence are not fully understood (7). The liver-specific miRNA, miR-122, positively modulates HCV infection through direct interaction with the 5’ UTR of the HCV genome and stimulation of HCV translation (8). Its overexpression is associated with inhibition of the IFN signaling pathway (9). Remarkably, reagents that can downregulate miR-122 have entered clinical development for HCV treatment (10).
Other miRNAs have also been reported to physically interact with the HCV genome and attenuate viral replication, namely let-7b, miR-196, miR-199, and miR-448 (11). In HCV-infected patients, lower expression levels of miR-29 have been observed in liver, while its overexpression inhibits viral RNA replication in HCV-infected hepatocytes (12). miR-130a expression has been up-regulated in liver biopsies from HCV-infected patients, as well as in HCV-infected hepatocytes (13).
HCV is a poor inducer pathogen of endogenous IFN. On HCV infection, the virus can modulate the function of certain cellular miRNAs involved in IFN production and the innate antiviral immune response, thereby exacerbating the infection (14). In addition, a number of reports have highlighted the importance of host cellular miRNAs in modulating responsiveness to exogenous IFN treatment (15). The deregulations of other miRNAs have been implicated in chronic HCV–associated inflammation, fibrosis, and cirrhosis, as well as the initiation and progression of liver cancer (16, 17).
2. Objectives
In this study, we attempted to identify special characteristics of the Egyptian HCV patients of predominantly genotype 4 by performing a selected 94 miRNA profile in 50 chronically HCV-infected patients. The correlation between individual miRNA expression profiles and clinicopathological variables was assessed for the sake of identifying potential biomarkers related to HCV-4 infection.
3. Patients and Methods
3.1. Patient and Clinical Samples
Fifty patients diagnosed with chronic HCV infection between 2011 and 2012 were enrolled in this study. All subjects were naive patients with no prior standard-of-care antiviral treatment. The study was reviewed and approved by the Cairo university hospital research ethics committee (REC), school of medicine, Cairo university. In addition, three normal biopsies were collected from liver transplantation donors. Written informed consent was obtained from all patients or their legal guardians.
Liver biopsies and blood samples were taken from each patient. Tissues were divided into two parts; one was used for histopathological examination, and the other was stored in RNAlater (Qiagen, Hilden, Germany) at -80°C until use. Liver biopsies obtained from normal liver transplantation donors were used as controls. For each biopsy sample, liver fibrosis and activity were scored according to the METAVIR classification system (18).
Patients were considered HCV positive if their serum tested positive in an enzyme-linked immunosorbent assay (ELISA; Detect-HCV 3, Adaltis, Milano, Italy) for HCV antibodies. All enrolled patients were HCV-RNA positive and the viral load was determined using quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) assay (Artus HCV RG RT-PCR, Qiagen). Patients who were co-infected with HBV or human immunodeficiency virus (HIV) and/or who had liver cirrhosis were excluded from this study. Liver function tests were conducted to determine patients’ levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin, albumin, alkaline phosphatase (Randox, Randox laboratories Ltd., London, UK) and α-fetoprotein (AFP; Adaltis). Bilharzial antibody titer was determined to assess the bilharzial status of the patients (NovaTec Immundiagnostica GmbH, Germany). HCV genotyping was assessed using the GEN-C reverse hybridization strip assay Kit (NLM diagnostic, Italy). All assays were carried out according to the manufacturers’ instructions.
3.2. miRNAs Extraction and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was isolated from 25 mg of liver tissue. Samples were first homogenized in QIAzol lysis reagent, then RNA was purified using miRNeasy® Mini Kit (Qiagen) following the manufacturer’s instructions. One microgram of RNA was used to prepare cDNA using miScript reverse transcription Kit (Qiagen) according to the supplied protocol. RNA concentrations were quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
The expression levels of 94 miRNAs were determined using miRNA primers. The selected miRNAs for this study were previously shown to be associated with liver health and chronic liver diseases. PCR reaction containing 1X SYBR green master mix (Qiagen), 200 nM of miRNA specific forward primer, and 200 nM of universal primers were performed in 96-well plates using 3 ng in 2.5 µL cDNA/well. The total reaction volume was 10 µL/well with initial denaturation at 95°C followed by 40 cycles at 94°C for 15 seconds, 55°C for 30 seconds, and 70°C for 30 seconds. The qRT-PCR experiments were performed on an ABI 7500 (Applied Biosystems, Foster city, CA, USA). The applied biosystems 7500 software was used to export the Ct values of different miRNAs to the Microsoft excel program. All samples were analyzed in duplicate.
3.3. Data Analysis
The relative expression of miRNAs was normalized using the mean expression value of all examined miRNAs in each sample as a normalization factor, following Mestdagh et al. (19). All Ct values above or equal to 35 were removed before calculating the mean of the remaining Ct values. The mean Ct value was calculated for each sample according to the Equation 1:

The ∆∆Ct and fold changes were calculated according to the Equation 2 (Fold Change = 2-ΔΔCt):

Heat-map and two-way clustering analysis were performed with a log2 fold of change using GENE-E software (Broad Institute, Inc.).
3.4. Statistical Analysis
SPSS version 17.0 for Windows (SPSS Inc. Chicago, IL, USA) was used for data management and data analysis. The chi-square test was used to test the observed distribution of miRNA as up- or down-regulated to an expected distribution. The non-parametric Mann–Whitney and Kruskal–Wallis tests were used to analyze differences in miRNA values according to categorical clinicopathological features. Spearman Rho correlation analysis was used to associate miRNA expression with numerical data. A P value < 0.05 was considered significant.
4. Results
Liver biopsy and blood samples were collected from 50 chronically infected HCV patients. As expected, 94% of the enrolled subjects in this study were genotype 4, and only 6% were of genotype 1. The levels of liver enzymes AST and ALT were mildly elevated. Eighty percent of the patients had a low–medium viral load. Liver biopsy samples revealed that most patients (90%) had mild to moderate fibrosis. Steatosis was mild in 78% and moderate in 12% of the samples (Table 1).
| Parameter | Value |
|---|---|
| Gendera | |
| Male | 36 (72) |
| Female | 14 (28) |
| Bilharziasisa | |
| Present | 32 (64) |
| Absent | 18 (36) |
| HCV titera (log Meq mL-1) | |
| Low (< 5.3) | 23 (46) |
| Medium (5.3 – 6) | 19 (38) |
| High (> 6) | 8 (16) |
| Activitya | |
| Minimal | 3 (6) |
| Mild | 13 (26) |
| Moderate | 32 (54) |
| Severe | 0 (0) |
| ND | 2 (4) |
| Stage of fibrosisa | |
| F0 | 0 (0) |
| F1 | 26 (52) |
| F2 | 15 (30) |
| F3 | 6 (12) |
| F4 | 1 (2) |
| ND | 2 (4) |
| Steatosisa | |
| Mild (< 33%) | 39 (78) |
| Moderate (33% - 66%) | 6 (12) |
| Marked (> 66%) | 3 (6) |
| ND | 2 (4) |
| Age, yb | 40.00 ± 11.99 |
| ASTb, IU/mL | 55.30 ± 37.34 |
| ALTb, IU/mL | 59.49 ± 37.87 |
| Alkaline phosphataseb, IU/L | 104.72 ± 40.85 |
| Albuminb, g dL-1 | 3.88 ± 0.83 |
| Bilirubinb, mg dL-1 | 0.92 ± 0.34 |
| Plateletsb, 103 µL-1 | 203.36 ± 57.16 |
Clinicopathological Features of the Hepatitis C Virus (HCV) Patients Enrolled in This Study
4.1. Profiling of 94 miRNA Expression Patterns in Hepatitis C Virus (HCV)-Infected Egyptian Patients
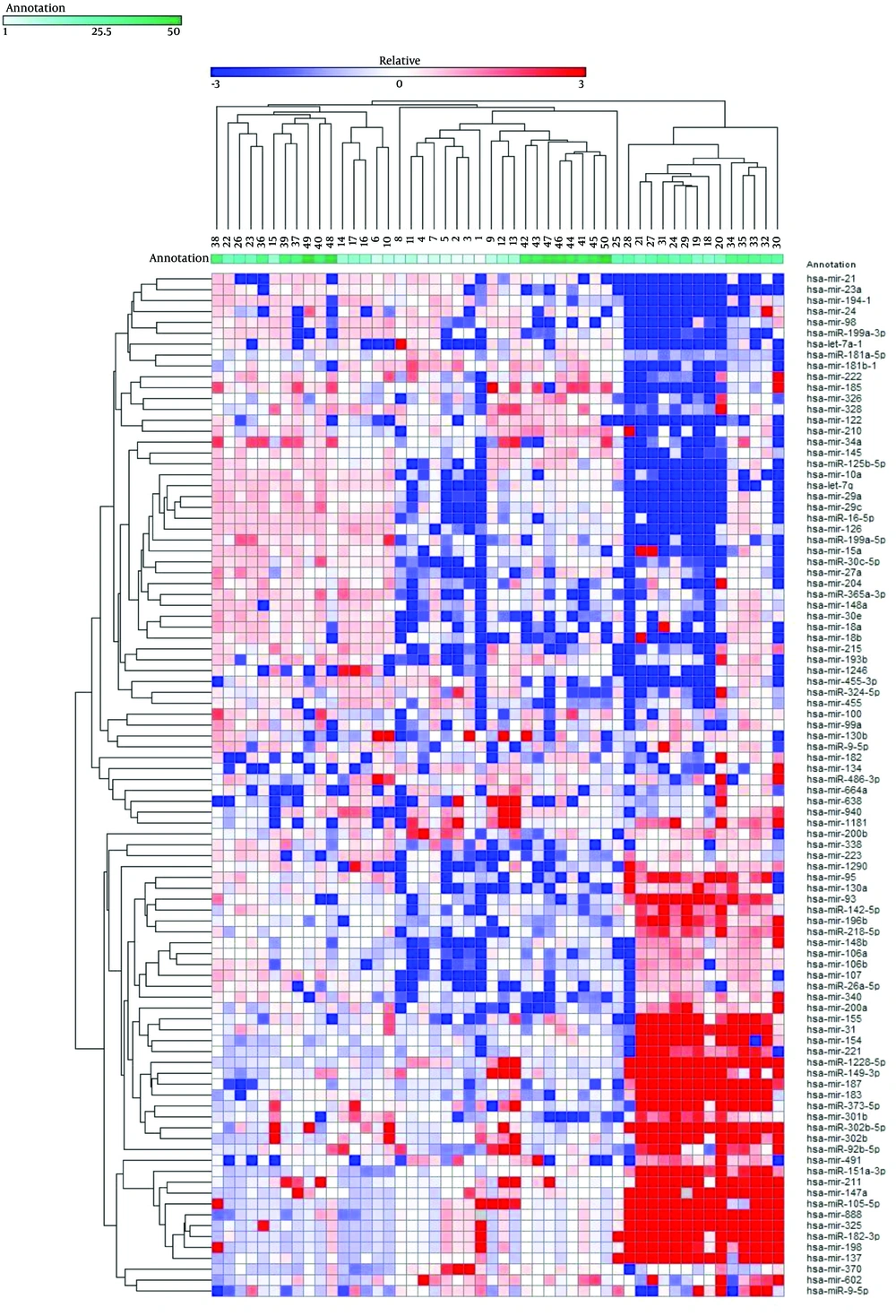
Expression levels of 94 mature miRNAs from 50 HCV liver biopsies were determined. The log2 fold of change was used to generate expression profile heatmap using Gene-E software. Two-way clustering analysis showed an association between the enrolled patients and the expression levels of the studied microRNAs (Figure 1). Forty-four out of the 94 analyzed miRNAs were significantly deregulated; 27 miRNAs exhibited at least a 4.0-fold increase, and 17 miRNAs exhibited at least a 4.0-fold decrease. To understand the role of the deregulated miRNAs in association with HCV infection, their published functions and proposed targets were summarized, as shown in Tables 2 and 3. A number of these miRNAs have been shown to be associated with HCV replication, pathogenesis, and associated liver complications.
The diagram represents 94 miRNAs’ expression profiling in 50 HCV patients. Each column represents a sample and each row shows the fold change of each miRNA in comparison to the average of normal samples. The differential regulation is indicated by red for up-regulated genes and blue for the down-regulated ones.
| miRNA | Average Fold Change | Functions Suggested in the Literature |
|---|---|---|
| let-7 | The let-7 members are involved in the regulation of STAT3, cell proliferation, and anti-inflammatory properties. | |
| let-7a | -7.6 | |
| let-7g | -5.6 | |
| miR-9 | -15.8 | Down-regulation is associated with liver fibrosis. |
| miR-10a | -6.3 | Down-regulated in hepatic stellate cells (HSCs). |
| miR-16 | -5.2 | Involved in the proliferation, clonogenicity, anchorage-independent growth, cell cycle (G1 phase arrest). |
| miR-18 | miR-18a/b is involved in the regulation of human embryonic stem cells (hESCs) and the epithelial–mesenchymal transition (EMT) of hepatocytes. | |
| miR-18a | -6.2 | |
| miR-18b | -7.9 | |
| miR-21 | -4.6 | Negatively regulates IFN signaling. It contributes to evasion of host immune system by targeting MyD88 and IRAK1. It regulates hepatocytes proliferation, and its expression level is correlated to liver fibrosis. |
| miR-24 | -13.3 | Overexpression increases proliferation; inhibits cell apoptosis in HCCs. |
| miR-30e | -4.1 | Can protect against chemokine ligand 4–induced liver fibrosis in a transforming growth factor-mediated pathway. |
| miR-99a | -4.9 | Down-regulated in HCCs. It suppresses HCC growth by inducing cell cycle arrest. |
| miR-122 | -4.9 | Involved in HCV replication and hepatic lipid metabolism. It acts as a tumor suppressor in hepatocarcinogenesis and is associated with poor HCV prognosis. |
| miR-126 | -4.5 | Involved in HCV infection. |
| miR-148a | -14.2 | Liver-specific microRNA. Promotes the hepato-specific phenotype and suppresses the invasiveness of transformed cells. |
| miR-194 | -17.2 | Overexpression abrogates the HCV infectivity of Huh7 cells. |
| miR-199a-3p | -8.7 | Inhibits HCV replication. It targets proliferation and migration/invasion signaling pathways in different cancers, especially HCC. |
| miR-223 | -7 | May affect inflammation, viral infection, and cancer development. |
List of Down-Regulated miRNAs in the Enrolled Hepatitis C Virus (HCV) Patients
| miRNA | Average Fold Change | Functions Suggested in the Literature |
|---|---|---|
| miR-31 | 250.1 | Up-regulated in chronic HCV patients’ urine samples. Up-regulated during T cell activation. Its expression is correlated with cirrhosis in HCC. |
| miR-92b-3p | 36.8 | miR-17-92 family genes are up-regulated, while miR-122 is down-regulated, in animals with HCC. |
| miR-95 | 4.6 | Promotes cell proliferation and targets sorting Nexin 1. |
| miR-105 | 7841.8 | Differential expression is related to antigen presentation and immune response during HCV infection. |
| miR-133b | 14.8 | Differential expression is related to proteasomes in HCV patients. |
| miR-137 | 1242.2 | Inhibits proliferation and invasion in different cell types. |
| miR-142-5p | 4.8 | miR-142-3p and miR-142-5p comprise two out of the seven most abundant miRNAs in T cells. |
| miR-147 | 541.3 | Associated with changes in the inflammatory capacity of immune cells by repressing tumor necrosis factor-alpha (TNF-α) and interleukin-6 (IL-6). |
| miR-149-3p | 21399.5 | May be involved in HCV entry, replication, and propagation. |
| miR-154 | 24.8 | Plays a role in proliferation and apoptosis. |
| miR-155 | 6.4 | miR-155 and miR-196b expression correlates with the detection of the antigenomic strand of HCV in peripheral blood mononuclear cells. |
| miR-183 | 58.6 | Up-regulation may be associated with onset and progression of HCC but not with patient survival. |
| miR-187 | 61.9 | IL-10–induced microRNA negatively regulates TNF-α, IL-6, and IL-12p40 production in Toll-like receptor-4 (TLR4)-stimulated monocytes. |
| miR-196b | 9 | miR-155 and miR196b expression correlates with the detection of the antigenomic strand of HCV in peripheral blood mononuclear cells. It can inhibit HCV replication when upregulated by INF-β. |
| miR-198 | 449.2 | Plays a role in cell proliferation. |
| miR-211 | 87.4 | Differential expression related to proteasomes in HCV patients. |
| miR-301b | 8.5 | Overexpressed in cervical and colon cancer. It could potentially increase Ras activation. |
| miR-302b | 151.3 | Regulates inflammatory response to bacteria by targeting TLR/IRAK4 circuits. Acts as a tumor suppressor by targeting EGFR and E2F3. |
| miR-302b-5p | 3990.9 | Specifically expressed in hESCs. |
| miR-325 | 184.3 | Contributes to the pathogenesis of preeclampsia. Involved in multicellular organism development and RNA splicing. |
| miR-328 | 8.7 | Increased in human primary biliary cirrhosis. Associated with MC liver and potentially regulates K-ras. |
| miR-373-5p | 28.7 | Up-regulated in hepatocytes infected with HCV; negatively regulates the type I IFN signaling pathway by suppressing JAK1 and IRF9. |
| miR-602 | 4.7 | Regulates tumor suppressive gene RASSF1A and is overexpressed in HBV-infected liver and HCC. |
| miR-888 | 151.7 | Overexpression inhibits RBL1 and SMAD4 in prostate cancer cells. |
| miR-1181 | 15.3 | Involved in HCV entry, replication, and propagation. |
| miR-1228-5p | 3026.7 | Prevents cellular apoptosis through targeting of MOAP1 protein. |
| miR-1290 | 4.2 | Overexpression affects hepatic differentiation in vitro. |
List of Up-Regulated miRNAs in the Enrolled Hepatitis C Virus (HCV) Patients
Remarkably, our data showed that the liver-specific miRNA-122 was strongly down-regulated in the majority of our patients, which contradicts several other previously published papers showing an increase in this miRNA correlated with its involvement in HCV infection and replication (20, 21). In contrast, our results were in agreement with Spaniel et al. who found that levels of miR-122 were reduced in Japanese patients with chronic hepatitis C (22).
4.2. The Correlation Between miRNA Expression Levels and Clinical and Bio-Chemical Data
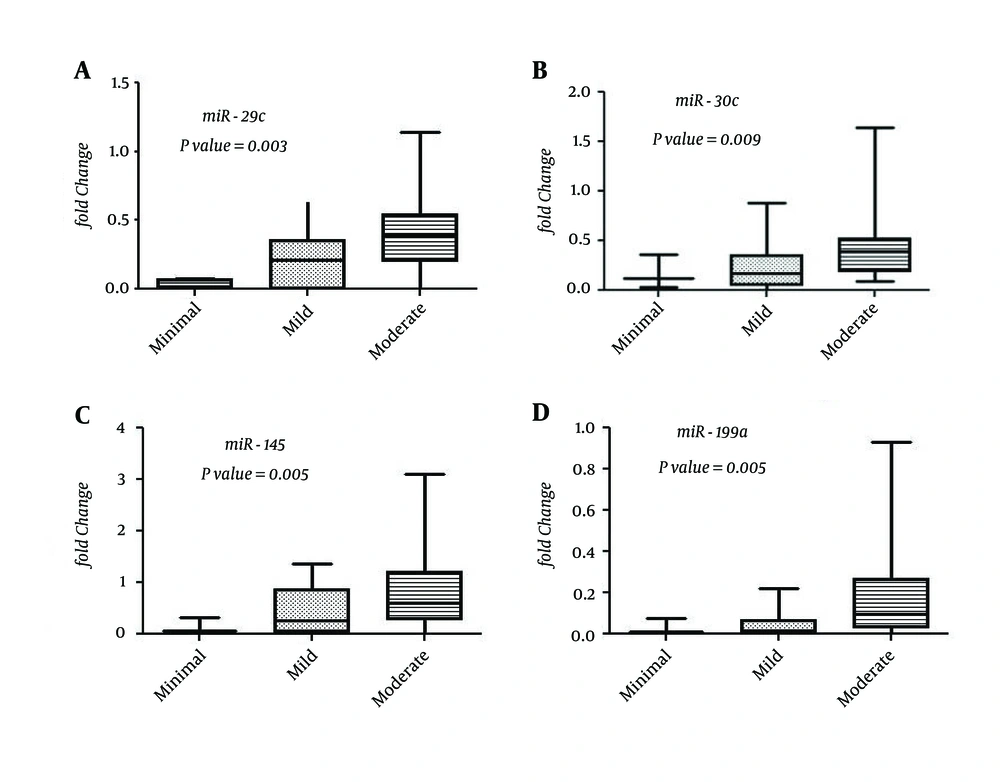
Expression levels of seven miRNAs (miR-29c, miR-30c, miR-126, miR-145, miR-199a, miR-199a-3p, and miR-222) tended to increase specifically in patients with an increasing grade of inflammatory activity (Figure 2). Expression of three other miRNAs (miR-95, miR-130a, and miR-142-5p) expression was inversely correlated with the serum albumin levels of the enrolled patients. No significant correlations were shown between the expression levels of analyzed miRNAs and basic clinical and biochemical characteristics of the studied patients, including age, viral load, serum liver enzymes, AFP level, presence of bilharziasis, or steatosis (data not shown).
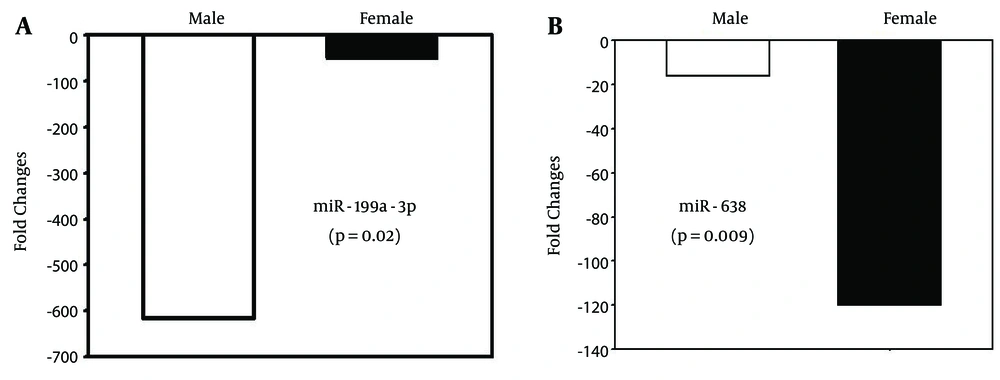
Our data analysis showed that overall expression of some miRNAs differed by patient gender. Five miRNAs, namely miR-21 (P = 0.001), miR-23a (P = 0.046), miR-126 (P = 0.045), miR-194 (P = 0.04), and miR-199a-3p (P = 0.02), showed down-regulation in male patients, while only one miRNA (miR-638) was significantly down-regulated in female patients (P = 0.009; Figure 3A and B).
5. Discussion
Over the last few years, several studies have examined the expression levels of certain miRNAs in association with hepatitis C viral infection. Some of the obtained data were provided in the context of HCC (23, 24). In this study, we aimed to identify changes in miRNA characteristics of naïve Egyptian patients with chronic HCV infection. We succeeded in identifying 44 miRNAs with at least 4.0-fold change compared to the normal controls. The identified host-altered miRNAs may contribute to the chronicity of HCV and mediated pathogenesis.
The combined samples clustering with the expression pattern of the studied 94 miRNAs could help to cluster the enrolled patients according to the progression of liver chronicity; further investigations are necessary. To develop a better understanding of the role of the deregulated miRNAs in association of chronic HCV infection, their suggested functions were manually curated from relevant published literatures (Tables 2 and 3).
The three liver-specific miRNAs analyzed in this study (miR-122, miR-148a, and miR-194) were significantly down-regulated in the majority of the patients. Our results are in agreement with a recent report regarding the reduced level of miR-122 in the tissues of chronic hepatitis C–infected patients and chimpanzees. The decrease of miR-122 in our study may have occurred after an initial rise at the beginning of the HCV infection, which was then followed by a decline, since all of our subjects were in the chronic phase of infection. This initial rise of hepatic miR-122 levels followed by a decline was shown to occur in chimpanzees (25). In recent studies, the hepatic miR-122 expression level has been shown to be reduced significantly with the severity of liver fibrosis in patients with chronic HCV infection (26, 27); this might fit with the clinical data of the enrolled patients in this study to some extent. The reduced level of miR-122 was found to be associated with a poor response to INF-based treatment (28). Thus, the selection of patients with chronic HCV infection for effective INF-based therapy may be achieved based on the hepatic expression level miR-122; further investigations on this topic are necessary.
Other differentially expressed miRNAs in this study have been shown to be associated with liver inflammation to fibrosis and HCC (let-7, miR-21, miR-223 (down-regulated), miR-105, miR147, miR-155, miR-187 (up-regulated) (29). Notably, the down-regulated miR-223 can act as a negative regulator of inflammation during viral infection. Its suppression is associated with chronic inflammatory response (30). miR-130a, a potential drug target in HCV treatment, was down-regulated in the examined hepatic samples (albeit not below the threshold level we set for this study). It has been reported that miR-130a overexpression can inhibit HCV replication by restoring host innate immune responses and/or down-regulating pro-HCV miR-122 (31). Restoring the expression level of miR-130a in infected patients may be a useful strategy to combat HCV infection.
We identified seven miRNAs (miR-29c, miR-30c, miR-126, miR-145, miR-199a, miR-199a-3p, and miR-222) that were significantly increased with the inflammation grade of the liver (Figure 2). Consistent with our data, it has been reported that members of miR-29 and miR-199 families correlate with the stage of liver fibrosis in HCV patients (32, 33). Identification of possible markers for the inflammation stage can be helpful in monitoring the disease progression and in avoiding the invasive biopsies used in these assessments.
Liver fibrosis is the most common final path of chronic liver diseases; this is influenced by the nuclear factor-kappa B (NF-κB) signaling pathway (34). miR-155, which was up-regulated in the present study, has been shown to have a negative effect on regulation of the NF-κB pathway through targeting different proteins, including myeloid differentiation primary response gene 88 (MyD88). Up-regulation of miR-155 was also reported in HCV-infected cells, leading to repression of the NF-κB signaling pathway (35). Meanwhile, miR-16, miR-199a, and miR-223, which were down-regulated in the present study, can regulate the NF-κB pathway by targeting key signaling protein genes (36, 37).
It was shown previously that c-Rel (a member of the NF-κB family) was able to negatively regulate miR-1228 by binding directly to its promoter site (38). In the present study, extremely high expression of miR-1228-5p was recorded (3,026.7-fold change) in liver cells infected with HCV. It would be interesting to study the effect of increased miR-1228 on the NF-κB pathway in infected cells.
Our results showed that a distinctive subset of miRNAs up-regulated simultaneously in some patients (miR-31, miR-105b, miR-147, miR-149-3p, miR-198, miR-302b-5p, and miR-1228-5p), in which they exhibited a greater than 200-fold change compared to normal subjects. Further studies are needed to reveal the significance of their expression level and the cellular machinery(ies) governing the miRNA-targeted relationship in chronically infected HCV patients.
It is interesting to note that a number of the deregulated miRNAs investigated in this study and their target genes, which are associated with advanced fibrosis or cirrhosis, play a critical role in the initiation and progression of HCC (Tables 2 and 3). In addition, three miRNAs, namely miRNA-18a, miR-18b, and miR-21, can promote cell proliferation and invasion and contribute to evasion of the host immune system. The role of several up-regulated miRNAs in association with chronic HCV infection is not yet fully understood (Table 3).
In conclusion, many of the dysregulated miRNAs in this study are associated with chronic liver disease and subsequent complications. This may reflect the potential mechanism(s) of persistent HCV infection to drive normal hepatocytes to malignancy via changing expression of several miRNAs, as a layer of gene expression control, in the host cell. Moreover, the identified expression profiles of some examined miRNAs might offer important points to consider for the treatment of naïve patients and management of chronically infected HCV patients in Egypt and around the world.