1. Background
Hepatitis B virus (HBV) infection is a major health problem worldwide (1, 2). The HBV genome is compact and consists of double-strand circular DNA of approximately 3.2 base pairs that encodes four partially overlapping open reading frames: surface (S), core (C), polymerase (P), and X genes. Ten genotypes (A to J) (3, 4) and more than 30 subgenotypes (5) of HBV have been identified based on the general rule (6). These genotypes arise during replication as a result of nucleotide misincorporation, in the absence of any proofreading capacity by viral polymerases (7-9).
Although mutations can occur randomly along the HBV genome, the overlapping open reading frames of HBV limit the number and location of variable mutants. Mutants have been described in all four genes of HBV, but have been more fully characterized in the pre-S, pre-core/core and polymerase regions (10). The hepatitis B surface antigen (HBsAg) protein is an important target of immune-mediated virus elimination. As a structural protein, HBsAg is an immune target. Selection pressure by HBs antibodies has led to the emergence of an immune escape mutation in this protein. As a result, it is no longer recognized by the host immune system and leads to occult hepatitis B infection (OBI) (11). A recent study of HBV infection showed that the rates of chronicity in patients infected during the perinatal period or childhood (30% - 90%) were higher than those infected in adulthood (12). Chronicity of virus causes the development of high immune responses leading to HBV escape mutants in patients with chronic infection (13).
After acute HBV infection, most adults appear to recover. The viral protein or DNA is typically not detectable in their blood, and they are generally not considered to be at risk for the disease. In other individuals, HBV infections persist. At least three distinct clinical states of viral persistence have been defined, based on serological findings in adults with chronic HBV infection: (1) a replicating phase, (2) a nonreplicating or low replicating phase, and (3) the more recently defined OBI. OBI is a form of long-term HBV infection. However, the clinical symptoms are undefined and differ from those of previously described forms of HBV (14). OBI is characterized by the presence of HBV DNA in the blood and liver in HBsAg-negative individuals, who may have antibodies to HBV core antigens (HBcAb) and HBsAg (HBsAb) (15).
A number of explanations have been proposed for the persistence of HBV DNA in HBsAg- negative samples. These include the integration of HBV DNA into the host’ chromosomes (16), genetic variations in the S gene (17), and the presence of immune complexes in which HBsAg may be hidden (18). In addition, OBI may be due to the window period after acute HBV infection, poor laboratory detection of HBsAg due to a low level of HBs antigenemia, underlying hepatitis C virus coinfection, immunosuppression, or other host-related factors (19).
Injecting drug users have a high risk of HBV infection because of hazardous behaviors, such as sharing needles and unsafe sexual activity, and they may be reinfected by other HBV strains during these risky behaviors (20). Thus, the rate of recombination different HBV genotypes and new HBV subtypes and mutations within the HBV genome can be expected to increase, leading to the emergence of undetectable HBsAg (i.e., OBI) (21). The likelihood of OBI infection is greater in long-term injection drug users because of the longer duration of sharing needles and other equipment. Another study reported that prison time (long term) and injection drug use were independently associated with HBV infection (22).
2. Objectives
There is limited evidence on the molecular characteristics of HBV among injecting drug users. This study was conducted to evaluate the basic molecular characteristics of the different forms of HBV associated with hepatitis and the associated risk factors in a well-known high-risk group (i.e., injecting drug users).
3. Patients and Methods
This cross-sectional study consisted of all IVDUs who had been referred to Loghman hospital in Tehran from January 2013 to January 2014. All the participants agreed to take part in the study after being informed about the study’s objectives. This study was approved by the ethics committee of Shahid Beheshti University of Medical Sciences (approval number 6-2015/9/6) and was in accordance with the Helsinki Declaration of 1964.
All the participants completed a questionnaire relating to demographics and risk factors. The sociodemographic information on the study group is provided in Table 1.
| Variable | OBI Negative | OBI Positive | P Value |
|---|---|---|---|
| Age, n = 169b | 0.465 | ||
| 20 - 30 | 24 (15.4) | 4 (30.8) | |
| 30 - 40 | 57 (36.5) | 5 (38.5) | |
| 40 - 50 | 46 (29.5) | 2 (15.4) | |
| > 50 | 29 (18.6) | 2 (15.4) | |
| Sex, n = 173 | 1 | ||
| Male | 157 (98.1) | 13 (100) | |
| Female | 3 (1.9) | 0 (0.0) | |
| Marital status, n = 170 | 0.201 | ||
| Married | 43 (27.4) | 6 (46.2) | |
| Single | 114 (72.6) | 7 (53.8) | |
| Employment status, n = 173 | 0.335 | ||
| Employed | 41 (25.6) | 5 (38.5) | |
| Unemployed | 119 (74.4) | 8 (61.5) | |
| Type of drug, n = 170 | 1 | ||
| Heroine | 36 (22.9) | 3 (23.1) | |
| Otherc | 46 (29.3) | 4 (30.8) | |
| Heroine + other | 75 (47.8) | 6 (46.2) | |
| Duration of drug usage, y, n = 147 | 0.437 | ||
| < 1 | 4 (3.0) | 0 (0.0) | |
| 1 - 5 | 19 (14.2) | 1 (7.7) | |
| 5 - 10 | 25 (18.7) | 5 (38.5) | |
| > 10 | 86 (64.2) | 7 (53.8) | |
| Frequency of injections in a day, n = 84 | 0.092 | ||
| 1 - 3 | 50 (67.6) | 5 (50.0) | |
| 3 - 6 | 19 (25.7) | 2 (20.0) | |
| > 6 | 5 (6.8) | 3 (30.0) | |
| History of imprisonment, n = 162 | 0.767 | ||
| Yes | 91 (60.7) | 8 (66.7) | |
| No | 59 (39.3) | 4 (33.3) | |
| History of sharing needle equipment, n = 82 | 0.037 | ||
| Yes | 13 (18.1) | 5 (50.0) | |
| No | 59 (81.9) | 5 (50.0) | |
| History of tattooing, n = 159 | 0.768 | ||
| Yes | 56 (38.4) | 4 (30.8) | |
| No | 90 (61.6) | 9 (69.2) | |
| History of surgery, n = 154 | 0.567 | ||
| Yes | 57 (40.4) | 4 (30.8) | |
| No | 84 (59.6) | 9 (69.2) | |
| History of dentistry, n = 106 | 0.744 | ||
| Yes | 61 (64.2) | 8 (72.8) | |
| No | 34 (35.8) | 3 (27.3) | |
| History of sexual behavior, n = 99 | 0.505 | ||
| Yes | 39 (43.3) | 5 (55.6) | |
| No | 51 (56.7) | 4 (44.4) | |
| HBV vaccination, n = 129 | 0.735 | ||
| Yes | 56 (46.7) | 5 (55.6) | |
| No | 64 (53.3) | 4 (44.4) | |
| History of blood transfusions, n = 85 | 0.214 | ||
| Yes | 20 (26.0) | 4 (50.0) | |
| No | 57 (74.0) | 4 (50.0) |
aValues are expressed as frequency (%).
bNumber of injection drug users who answered each question, n = 229.
cOther drugs, including crack and crystal.
A blood sample of 5 mL was collected from each participant, and the serum was separated by centrifuge and placed in sterile serum storage vials, stored at -70°C until tested.
3.1. Serological Tests
The presence of total HBcAb in the samples was tested using an enzyme-linked immunosorbent assay (ELISA) and a commercial kit for HBcAb detection (Diapro, Milan, Italy). The presence of the HBsAg marker in HBcAb-positive samples was tested using a commercial kit for HBsAg detection (Diapro, Milan, Italy).
3.2. DNA Extraction, Nested Polymerase Chain Reaction, and Gel Electrophoresis
HBV DNA was extracted from a 200 µL aliquot of HBcAb-positive sera (either HBsAg- positive or HBsAg-negative) using a high pure viral nucleic acid kit (Roche, Germany), according to the manufacturer’s instructions. To investigate the HBV genotypes, the partial HBV S gene was chosen for amplification, with specified primers. The nested polymerase chain reaction was performed for amplification of this segment. In the first round, the polymerase chain reaction consisted of the following program: 94°C for 5 minutes, followed by 35 cycles at 94°C for 30 seconds, 56°C and 72°C for 1 minute, and 72°C for 10 minutes. A similar program was applied in the second round of the polymerase chain reaction but with a different annealing temperature (62°C for 30 seconds).
The first polymerase chain reaction was performed using the sense primer S1 (5′- CCTGCTGGTGGCTCCAGTTC- 3′, Sense, nt 56 - 75) and the antisense primer S2 (5′- CCACAATTCKTTGACATACTTTCCA- 3′, Antisense, nt 979 - 1003) to yield a 1000 bp amplicon of the HBV genome. The second polymerase chain reaction was performed using the sense primer S6 (5′- GCACACGGAATTCCGAGGACTGGGGACCCTG- 3′, Sence, nt 133 - 146) and the antisense primer (5′- GACACCAAGCTTGGTTAGGGTTTAAATGTATACC- 3′ Antisense, nt 823 - 857 ) to yield a 700 base pair amplicon of HBV genome (23). The amplified products were subjected to electrophoresis in 1.5% agarose gel and evaluated under ultraviolet translumination. The specified amplified HBV DNA product was determined by comparing with the 100 bp DNA ladder (Fermentas, USA), which was used as a DNA size marker.
3.3. DNA Sequencing and Phylogenetic Analysis
The HBV genotypes and subtypes were determined by a phylogenic analysis based on the partial sequence of the HBV surface antigen (687 bp). A direct sequencing protocol was used, with the nested polymerase chain reaction protocol targeting HBsAg. To generate a phylogenetic tree, the HBV sequences were compared to those of an A-H defined HBV strain retrieved from GenBank. BioEdit software, version 7.0, was used to align the sequences. The phylogenetic tree was constructed by the neighbor-joining method. Bootstrap re-sampling and reconstruction were carried out 1,000 times to confirm the validity of the phylogenetic tree. MEGA 5.0 software was used for the phylogenetic and evolutionary analysis. Subtypes were deduced from the sequence of the viral genome region by encoding the HBsAg (24).
3.4. Statistical Analysis
Fisher’s exact test and the statistical package for social sciences (SPSS), version 20, were used to analyze the data. A P value of less than 0.05 was considered statistically significant. To avoid interruptions in the statistical analysis, injecting drug users who did not answer the questions were excluded from the data analysis.
4. Results
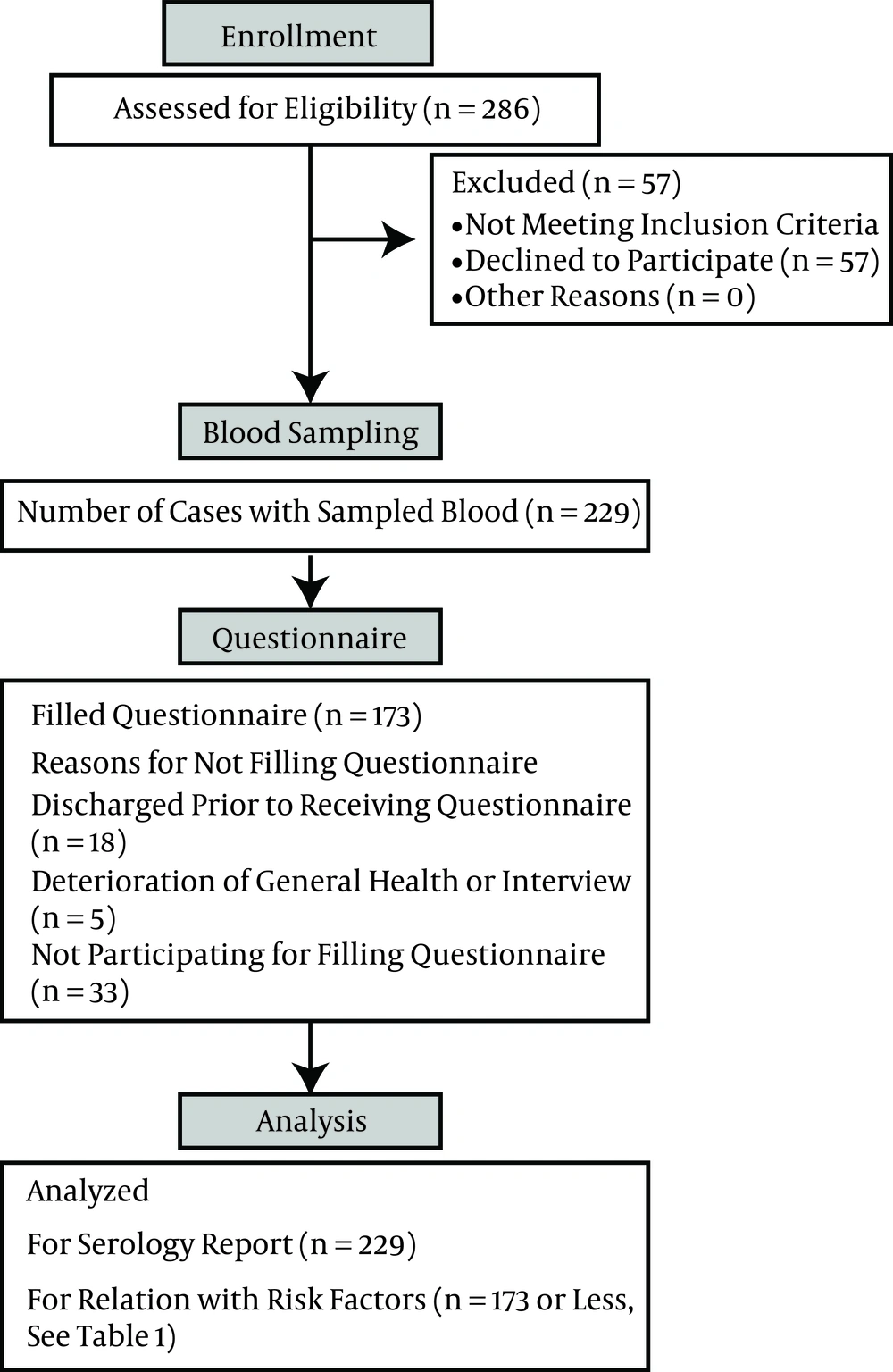
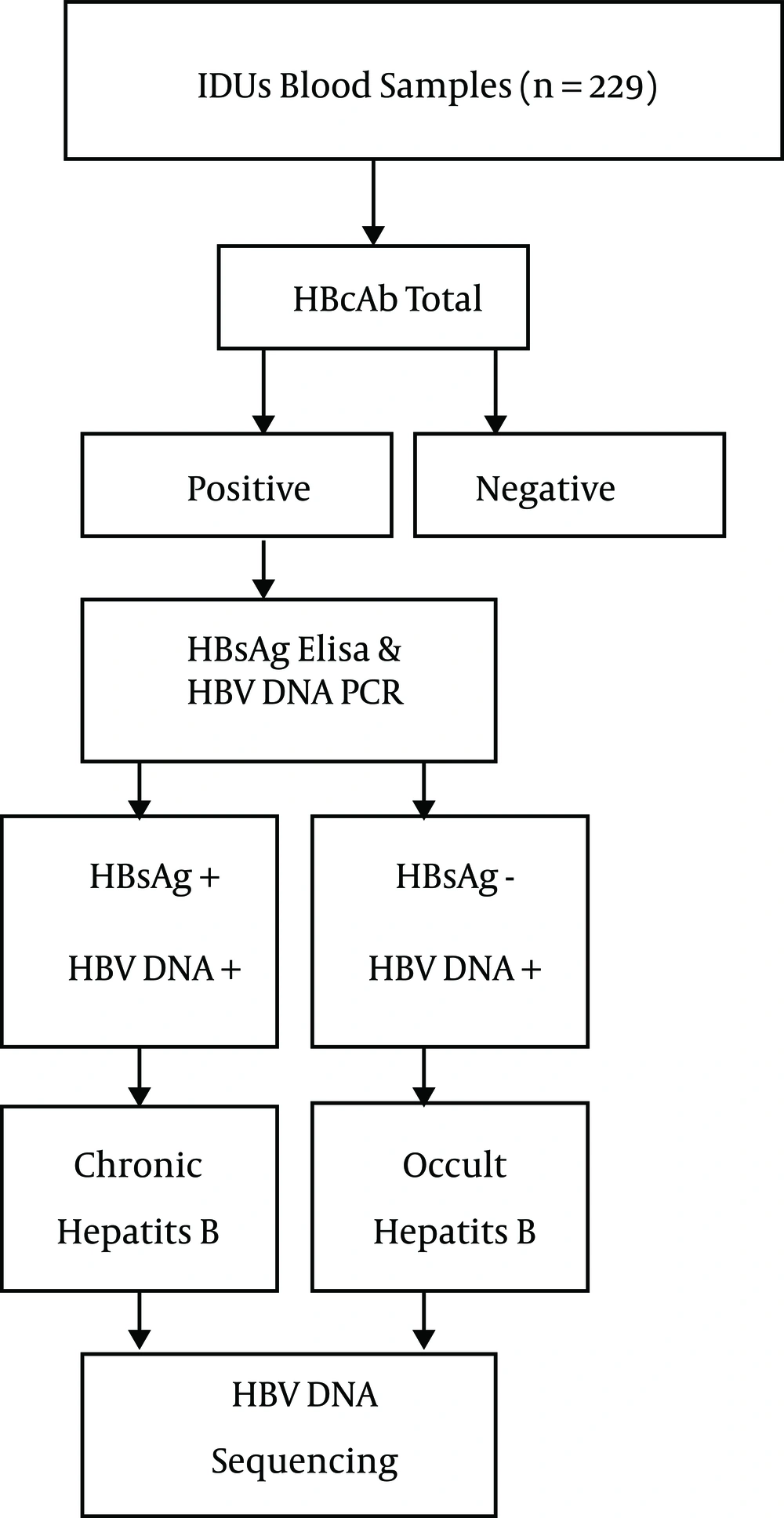
The algorithm of the patient flow for blood sampling and completion of the questionnaire are depicted in Figure 1. The algorithm for blood tests and HBV DNA sequencing is outlined in Figure 2. Of 229 IVDU serum samples, 64 (28%) were HBcAb positive. Of these HBcAb- positive samples, five were HBsAg positive, and 59 were negative for this marker. The highest rate of HBsAg positivity was among those who had more than a 10-year record of drug use and a history of imprisonment.
The presence of HBV DNA was analyzed in two groups of samples: HBsAg-/HBcAb+ (n = 59 samples) and HBsAg+/HBcAb+ (n = 5 samples). HBV DNA was successfully amplified in three cases of HBsAg+/HBcAb+ (chronic carriers) and 13 cases of HBsAg-/HBcAb+ (OBI carriers). Table 1 shows the association of OBI with the demographic variables and risk factors associated with IVDU behavior. According to the results, only sharing a syringe was associated with OBI (P = 0.037). There were no significant associations between the demographic variables and other risk factors for OBI (Table 1).
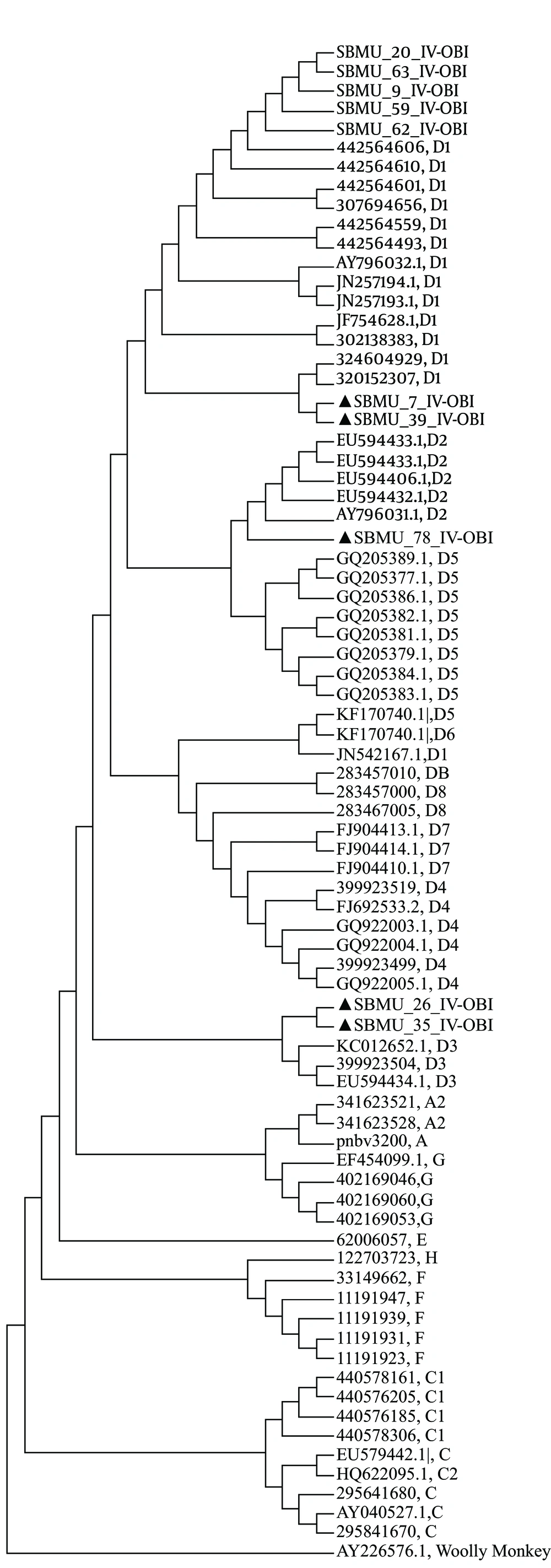
In total, 16 sequences with unambiguous sequencing chromatographs were subjected to a phylogenetic analysis. The phylogenic tree of the two aforementioned groups was constructed separately to evaluate possible between-group differences. The results of the phylogenetic analysis revealed genotype D among all the samples. Nine samples were subgenotype D1, two were subgenotype D, and two were sub-genotype D3. The HBV subtype was deduced from the amino acid of the HBV S gene (Figure 3 and Table 2).
| Samples | Genotype | Subgenotype | Subtype | Nucleotide Substitution | Missense Mutation | Amino Acid Change |
|---|---|---|---|---|---|---|
| OBI cases | D | D1, D2, D3 | ayw2 | T22C, T23G, T26A, G36T, T44A, C136G, T206A, C219T, G227A, G233T, T245C, T247C, A302G, A327G, T339C, T340C, C358A, G362A, T378A, C379A, C407A, G440A, T473A, G475C, G492A, G502A, G529A, G550C, G553A, T569G, T612A, T616G, C640T, T658C, T663G | T22C, A610C, G529A | F8L, S204R, V177M |
5. Discussion
Molecular and epidemiological studies of HBV are important to formulate HBV prevention strategies. The prevalence of HBV infection varies, according to various risk factors, socioeconomic, and the initial burden of infectious markers in the community, which vary within different regions of a country (25). According to the results of this study, 2.1% of IVDUs in Tehran were HBsAg positive, and 28% of IVDUs in Tehran were HBcAb positive. The rate of HBsAg positivity in this study is in accordance with that found in our previous study (26). It is lower than that reported in studies performed in other areas of Iran, possibly due to HBV vaccination. Since HBV vaccination was added to the Iranian national program of immunization, the prevalence of HBV has decreased throughout the country. The lower rate may also be explained by a harm-reduction intervention, which was introduced in 2005 in Iran (27).
A previous study in Iran reported that the prevalence of HBsAg was 2.6% and that the prevalence of HBcAb was 2.6% in the general population (28), which was lower than that found in the current study. In comparison with other Iranian studies, the rate of HBcAb positivity was higher in the present study. This may be explained by past infection. Based on this finding, it cannot be concluded that HBV infection has decreased among IVDUs. Further investigation is needed, such as evaluating HBsAb, to shed more light on this topic.
In the present study, the overall prevalence of OBI in IVDUs was 5.6%. Another study in Iran of 153 IVDUs reported a HBcAb prevalence of 7.2%, although HBV DNA was not detected in any of the HBcAb-positive cases, as the study did not include the real time polymerase chain reaction (29). In a study of the rate of OBI in Iranian HIV-positive patients with isolated HBcAb, HBV DNA was detected in 13.6% of the patients (14). The same study showed that OBI was relatively common in HIV-infected patients with isolated HBcAb, regardless of age, sex, aminotransferase levels, and treatment with antiretroviral drugs. The relatively high rate of OBI in that population was based on computing the percentage of OBI-positive individuals among HBcAb-positive patients. The rate of OBI in the total selected population in the study was 2.8% (3 of 106 patients), which was lower then the rate in the present study (2.8% versus 5.6%). On the other hand, the method used to determine the positivity of HBV DNA was the real time polymerase chain reaction, which is less assuring compared to the nested polymerase chain reaction method used in our study. As noted elsewhere, the sensitivity of the HBV DNA assay and that of other molecular biology techniques, in addition to the sample size and composition of the study population, can affect the rate of OBI detection (30).
The results showed no significant association between the demographic parameters of the participants and risk factors with the rate of OBI, except sharing a needle. Of note, however, all the OBI-positive cases were men, mainly aged between 30 and 40 years. Most of these were also unemployed, with a history of imprisonment and multiple sexual partners or high-risk sexual behaviors. According to the findings, the risk of infection increases in accordance with the length of time that someone has been an injection drug user. In some reports from Iran and other countries regarding imprisonment, the frequency and duration of drug injection were demonstrated to be risk factors for HBV infection (26). Although a number of studies have investigated HBV infection and associated risk factors, none have focused on OBI. The results of the present study on demographic and risk factors associated with IVDUs and the rate of OBI are partly consistent with a study in Taiwan, which showed that the prevalence of occult HBV infection was positively correlated with increasing 10-year categories of age (31). According to that study, older injecting drug users may have a longer history of drug use, which reflects an effect of cumulative exposure.
The present study found a predominance of genotype D in the injecting drug users. Interestingly, a previous study identified a significant association between genotype D and injecting drug use (32). The predominance of genotype D observed in the present study is consistent with findings reported elsewhere. In the present study, the phylogenetic analysis revealed genotype D as the current genotype in injecting drug users; it also showed subgenotypes D1 and D2 and D3 with subtype ayw2 in this high-risk group.
Previous molecular epidemiological studies of the HBV S and C regions of Iranian patients with chronic HBV disease (33, 34) and five complete genome sequences (35) demonstrated that genotype D was the only HBV genotype in the Iranian population. One study showed that the majority of Iranian strains were subgenotype D1 and subtype ayw2, with small numbers of D2/ayw3 and D3/ayw2 isolates also detected (36). Another study demonstrated that all the strains tested belonged to genotype D, subgenotype D1, and subtype ayw2, although two strains with subtype ayw3 and adw2 were observed (13). Similarly, in a study of inmates in Iran, genotype D1 was the only predominant genotype (37). According to that study, genotype D1 was mostly transmitted by intravenous drug use, and a direct relationship was observed between genotype D and injection drug use. The same study reported that injection drug use was responsible for the HBV genotype D epidemic in Iran.
In the present study, the genetic variability analysis of Iranian injecting drug users showed a number of mutations and substitutions within the sequence of the S region (Table 2). In the study, of 10 amino acid mutations, five immune escape mutants were identified in injecting drug users with chronic disease. These mutations were outside the a-determinant region. Various point mutations in the sequence of HBsAg have been found among Iranian HBV isolates (36). Two studies investigated different amino acid mutations in immune epitopes, such as B cells, T- helper cells, and cytotoxic T lymphocytes (38, 39). A study on HBV surface protein mutations among Iranian high-risk patients with OBI resulted in nonfunctional HBsAg, in which the insertion of a single nucleotide in two cases caused a frame shift and single nucleotide replacement, and premature stop codons at Leu15 and Gly10 in the other two cases. An amino acid substitution at amino acid position 207(S207N) was found in the other isolates. Hamkar et al. suggested that “a” region mutations did not play a major role in HBsAg detection and that other genetic and nongenetic factors may be responsible for the failure to detect HBsAg in routine laboratory tests (40).
The low rate of genomic variations, such as genotypes, subtypes, and mutations, in the high-risk group in the present study is interesting. It may be explained by i) the low rate of chronic HBV infections among the injecting drug users and ii) the acquisition of infection during adulthood. The latter may lead to less interaction of the virus with the immune system and therefore a low rate of diversity in the HBV genome, especially in immune-escaped regions. According to Shahmoradi et al., the progression of the chronicity of HBV as a result of mutations can be found in chronic HBV patients (38). Vaezjalali et al. demonstrated that the frequency of a stop codon mutation in the S region and basal core promoter in strains derived from cirrhotic patients was higher than among strains derived from asymptomatic carriers. Other studies conducted in Turkey and Korea reported the same results (13). The chronicity of the virus causes the development of high immune responses, leading to HBV escape mutants in patients (13). It can be said that the accumulation of variation and mutations needs a long period of chronicity.
This study provided clear insight into the rate of OBI among IVDUs in the capital of Iran and revealed that genotype D was prevalent in this high-risk group. Other variations, genotypes, and subtypes were also detected among the infected injecting drug users. The results highlight the prevalence of OBI infection in IVDUs and demonstrate that they are a potential source of HBV transmission because of sexual, parenteral, and other risk factors associated with HBV infection. The findings suggest that additional attention should be given to improving the quality of informing people and easy access to health care information in our society about the possible transmission of HBV through the individuals without any symptoms. Furthermore, they point to the need to extend harm-reduction initiatives and strategies, such as vaccination, to prevent HBV transmission among IVDUs in Iran. Further investigations, such as cloning of derived strains (in order to assess to virus population), are needed to unravel the molecular characterization of OBI.