1. Background
According to the report of world health organization (WHO), more than 130 - 150 million people are infected with hepatitis C virus (HCV) and approximately 500,000 HCV-related deaths occurred annually (1). Chronic infection with HCV may lead to liver diseases, such as cirrhosis and hepatocellular carcinoma (HCC) (2, 3). In the recent decade, standard of care for treatment of HCV infection was combination therapy with pegylated-interferon (PEG-IFN) plus ribavirin (RBV). During the treatment with this regimen, patients must be monitored in order to manage unwanted side-effects. Bone marrow suppression and hemolytic anemia which may respectively occurred by PEG-IFN and RBV are the major side-effects of combination therapy. These side-effects of treatment may lead physicians for modifications or in some cases termination of therapy. Eventually, this issue may affect the final goal of HCV treatment, which is the eradication of infection (4-6).
It has been found that both virus and host parameters play a great role in HCV natural history (7). Recently, two functional single nucleotide polymorphisms (SNPs) including rs1127354 and rs7270101 within inosine triphosphatase (ITPA) gene were found to influence RBV-induced hemoglobin (Hb)-decline. Some studies showed that these SNPs were associated with platelet (Plt)-decline as well (8, 9). Another SNP which is associated with ITPA functional gene polymorphisms is rs6051702 within C20orf194 gene. These polymorphisms are appropriate candidates for predicting the RBV-induced Hb-decline (8-11).
2. Objectives
This study aimed to evaluate the effect of host genetic factors including ITPA rs1127354 and rs7270101 and C20orf194 rs6051702 polymorphisms and other baseline characteristics of patients on hematological changes including Hb-, Plt- and white blood cell (WBC)-decline at week 4 of treatment with PEG-IFN plus RBV in chronic hepatitis C (CHC) patients.
3. Patients and Methods
3.1. Study Population
In this cross-sectional study , 168 treatment-naive HCV-infected patients who had HCV RNA > 50 IU/mL for more than 6 months were evaluated and treated at Tehran Blood Transfusion Hepatitis Clinic (affiliated to Iranian Blood Transfusion Organization (IBTO)) and Tehran Hepatitis Clinic (affiliated to Baqiyatallah Research Center for Gastroenterology and Liver Diseases (BRCGL)) during 2011 - 2015. The patients who were coinfected with hepatitis B virus or human immunodeficiency virus, decompensated cirrhotic patients (based on histologic or imaging or clinical findings), cases with HCC, patients with creatinine clearance less than 50%, and who had contraindication for interferon therapy including severe heart failure, poorly controlled diabetes mellitus, poorly controlled psychiatric disorder, and also patients with baseline Plt count below 50,000/mm3 and/or WBC count below 2,000/mm3 were excluded from this study. In this study, male patients with baseline Hb lower than 13 g/dL and female patients with baseline Hb lower than 12 g/dL were excluded. All study participants provided informed consent and the study design was approved by the ethics committee of BRCGL. The study protocol conforms to the ethical guidelines of the 1975 declaration of Helsinki.
3.2. Treatment Regimen and Definition of Clinical Endpoints
All of the patients were treated with combination of PEG-IFN-α-2a (Pegasys, Roche, Basel, Switzerland) and RBV (Copegus, Roche, Basel, Switzerland) or PEG-IFN-α-2b (Pegintron, Schering-plough, Puerto Rico, USA) and RBV (Rebetol, Schering-plough, Puerto Rico, USA). The dose of Pegasys was 180 μg subcutaneously once a week in combination with oral RBV 800 – 1,200 mg per day according to patients’ weight. The dose of Pegintron was 80, 100 or 120 μg subcutaneously once a week according to patients’ weight in combination with oral RBV 800-1,200 mg per day according to patients’ weight.
The decline of hematological components at week 4 of therapy was chosen as the clinical endpoint. Several cut-off values were assessed for each blood component and the best cut-off value according to the lower P value for each parameter was taken.
3.3. Laboratory Assessments
The HCV RNA level was assessed using the COBAS® TaqMan® HCV test version 2.0 (Roche Diagnostics) according to manufacturer’s instructions. In this study, rs1127354, rs7270101 and rs6051702 SNPs were assessed using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. Briefly, genomic DNA was extracted from patients’ peripheral blood using the QIAmp DNA blood mini kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. The PCR were performed using Accupower PCR PreMix (Bioneer Corp., Daejeon, South Korea) with the following conditions: 94°C for 5 minutes; 35 cycles of 94°C for 20 seconds, 58°C (rs1127354/ rs7270101) or 54°C (rs6051702) for 20 seconds and 72°C for 20 seconds, followed by 72 °C for 5 minutes. Primers used for the reaction included: rs1127354/rs7270101, 5'-AGATGGGCAGCAGAGTTATCG-3' and 5'-AAGACAGAGAAATCCAACCATCTTTTAAGAA-3'; or rs6051702, 5'-CAAAAACTCACCATATAACAGGGGTTATGC-3' and 5'-TGGGGTGGTGAAGTGTGGTAAGTT-3'. The PCR amplicons were then digested for ≥ 1 hour with 10 U of restriction endonulease: XceI for rs1127354 or MboII for rs7270101 or Mph1103I for rs6051702 (Fermentas of Thermo Fisher Scientific, Waltham, MA, United States). The digested PCR products were separated on 3% agarose gel revealing the following sized fragments: rs1127354, 135 and 78 bp for the AA genotype, 213, 135 and 78 bp for the CA genotype or 213 bp for the CC genotype; rs7270101, 173 and 40 bp for the CC genotype, 213, 173 and 40 bp for the AC genotype or 213 bp for the AA genotype; rs6051702, 230 and 31 bp for the AA genotype, 261, 230 and 31 bp for the AC genotype or 261 bp for the CC genotype.
3.4. Statistical Analysis
Categorical variables were expressed by frequency and percentage. Continuous variables with normal distribution were expressed by mean ± standard deviation (SD) and continuous variables deviated from normal distribution by median (interquartile range). Fisher-exact test was used for analysis of categorical variables, t-test for continuous variables with normal distribution, and Mann-Whitney U-test for continuous variables deviated from normal distribution. In order to analyze the genetic associations, the additive genetic models were chosen including rs1127354 CC vs. CA + AA, rs7270101 AA vs. AC + CC and rs6051702 AA vs. AC + CC. Hardy-Weinberg equilibrium (HWE) was assessed for rs1127354, rs7270101 and rs6051702 SNPs and the linkage disequilibrium (LD) between these SNPs was calculated by the Haploview software. All baseline variables with a P value less than 0.2 in univariate analysis entered to a logistic regression model. P value less than 0.05 was considered to be statistically significant. Statistical analysis was performed using SPSS version 23 (SPSS Inc., Chicago, Illinois, USA). Statistical graphs were generated using GraphPad Prism version 6.
4. Results
4.1. Patients’ Characteristics
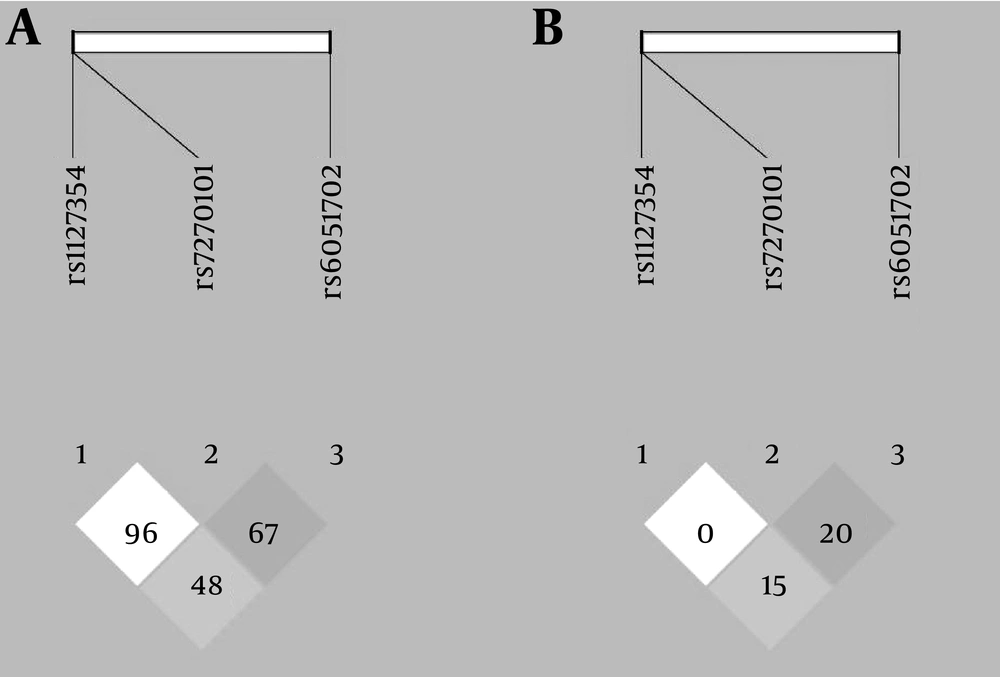
A total of 168 CHC patients under treatment of PEG-IFN plus RBV were determined. Most of the patients were male and also more than half of the patients were infected with HCV genotype-1. Baseline characteristics of the study population and also the genotypic distribution of the SNPs are shown in Table 1. The distribution of rs1127354 and rs7270101 genotypes were in HWE; however, the distribution of rs6051702 genotypes was deviated from HWE (P = 0.812, P = 0.319 and P = 0.006, respectively). The LD analysis plot for D’ and r2 are shown in Figure 1. The LD between rs1127354 and rs7270101 was not observed (D’ = 0.96, r2 = 0.0). The LD between rs6051702 with rs1127354 and rs7270101 are low (D’ = 0.48, r2 = 0.15 and D’ = 0.67, r2 = 0.2, respectively).
| Baseline Characteristics of Patients | Values |
|---|---|
| Gender | |
| Male | 154 (91.7) |
| Female | 14 (8.3) |
| Age, y | |
| Median (IQR) | 41 (17.0) |
| Range (min - max) | (23.0 - 77.0) |
| BMI, kg/m2b | |
| Mean ± SD | 26.66 ± 4.08 |
| Range (min - max) | (18.8 - 37.1) |
| Baseline Hb, g/dL | |
| Mean ± SD | 15.87 ± 1.41 |
| Range (min - max) | (12.2 - 19.4) |
| Baseline WBC, cell/mm3c | |
| Mean ± SD | 6840 ± 1780 |
| Range (min - max) | (2300 - 12800) |
| Baseline Plt, cell/mm3c | |
| Mean ± SD | 205922 ± 64141 |
| Range (min - max) | (56000 - 450000) |
| Baseline ALT, IU/Lc | |
| Median (IQR) | 67.0 (75.0) |
| Range (min - max) | (12.0 - 313.0) |
| Baseline AST, IU/Lc | |
| Median (IQR) | 41.0 (35.0) |
| Range (min - max) | (16.0 - 232.0) |
| HCV RNA, log IU/mLc | |
| Median (IQR) | 6.0 (6.56) |
| Range (min - max) | (3.01 - 7.52) |
| HCV genotypes | |
| 1a | 78 (46.4) |
| 1b | 9 (5.4) |
| 1 d | 7 (4.2) |
| 3a | 74 (44.0) |
| rs1127354 genotype | |
| CC | 136 (80.95) |
| CA | 30 (17.85) |
| AA | 2 (1.20) |
| MAF, % | 10.1 |
| rs7270101 genotype | |
| AA | 144 (85.7) |
| AC | 24 (14.3) |
| MAF, % | 7.1 |
| rs6051702 genotype | |
| AA | 127 (75.6) |
| AC | 33 (19.6) |
| CC | 8 (4.8) |
| MAF, % | 14.6 |
aValues are expressed as No. (%) unless otherwise indicated.
bThe data were missed in more than 10% of the patients.
cThe data were missed in less than 10% of the patients.
dThe patients with undetermined HCV genotype-1 subtype
4.2. rs1127354, rs7270101, rs6051702 and Hematological Changes at Week 4 of Treatment With Pegylated-Interferon Plus Ribavirin
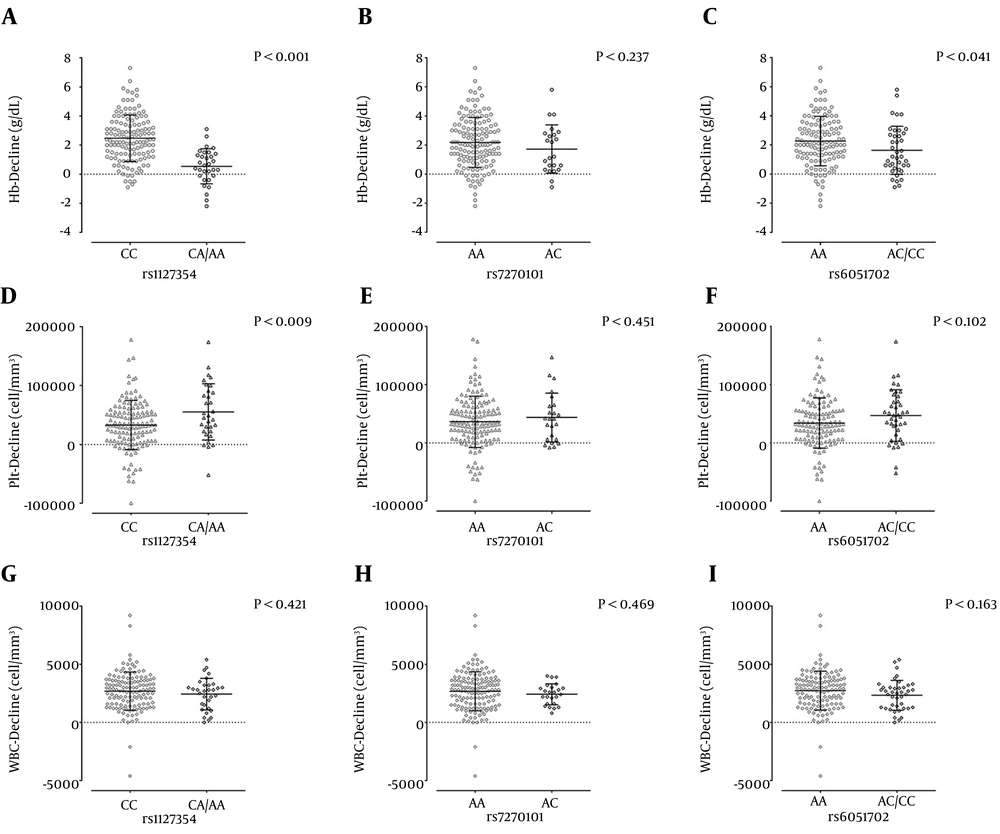
The effect of rs1127354, rs7270101 and rs6051702 polymorphisms on the hematological changes are shown in Figure 2. As it is obvious the Hb-decline at week 4 of treatment was affected by the rs1127354 and rs6051702. There was no significant association between rs7270101 and Hb-decline. Platelet-decline was affected by rs1127354; however, no significant association was observed between rs7270101 and rs6051702 with Plt-decline. In the other words, the carriers of A allele of rs1127354 in contrast to patients with the CC genotype had more Plt-decline. There was no association between these SNPs and WBC-decline at week 4 of treatment. After finding that these SNPs may just affect the Hb- and Plt-decline, these two parameters were assessed by univariate and multivariate analysis in order to determine the factors that may affect the Hb- and Plt-decline at week 4 of treatment. According to the best cut-off values, patients were classified into two groups for each hematological parameter; patients with ≤ 2 or > 2 g/dL Hb-decline and ≤ 80,000 or > 80,000 cell/mm3 Plt-decline.
A – C, rs1127354, rs7270101 and rs6051702 (respectively) and Hb-decline at week 4 of the treatment; D –F, rs1127354, rs7270101 and rs6051702 (respectively) and Plt-decline at week 4 of the treatment; G – I, rs1127354, rs7270101 and rs6051702 (respectively) and WBC-decline at week 4 of the treatment, The lines and bars are representative of mean and standard deviation. P values were obtained by t-test.
4.3. Evaluation of Factors Affecting Hemoglobin-Decline at Week 4 of Treatment With Pegylated-Interferon Plus Ribavirin
In univariate analysis, no association was found between the age, sex, BMI, ALT, AST, HCV RNA level, rs7270101, rs6051702 and RBV dose with Hb-decline at week 4 of the treatment (P > 0.05) (Table 2). However, HCV genotype-3 (P = 0.008, OR = 0.43) and rs1127354 CA + AA (P < 0.001, OR = 0.05) were found to prevent Hb-decline at week 4 of the treatment (Table 2). Multivariate analysis confirmed this finding and it was found that HCV genotype-3 (P = 0.003, OR = 0.36) and rs1127354 CA + AA (P < 0.001, OR = 0.04) could prevent the Hb-decline at week 4 of the treatment (Table 2).
| Variables | Hb-Decline | Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|---|---|
| ≤ 2 (n = 87) | > 2 (n = 81) | OR (95% CI) | P Value | Adjusted OR (95% CI) | Adjusted P Value | |
| Age, median (IQR) | 39 (18.0) | 40 (17.0) | - | 0.311 b | - | - |
| Gender | - | - | - | > 0.999 c | - | - |
| Male | 80 (92.0) | 74 (91.4) | Ref. | - | - | - |
| Female | 7 (8.0) | 7 (8.6) | 1.08 (0.36 - 3.22) | - | - | - |
| BMI, median (IQR) | 26.8 (5.6) | 26.5 (6.2) | - | 0.942 b | - | - |
| ALT, median (IQR), IU/L | 73 (93.0) | 68 (63.0) | - | 0.243 b | - | - |
| AST, median (IQR), IU/L | 45 (40.0) | 40 (24.0) | - | 0.289 b | - | - |
| HCV RNA level, IU/mL | - | - | - | 0.626 c | - | - |
| > 600,000 | 51 (60.7) | 50 (64.9) | Ref. | - | - | |
| < 600,000 | 33 (39.3) | 27 (35.1) | 0.83 (0.44 - 1.59) | - | - | - |
| HCV genotype | - | - | - | 0.008 c | - | 0.003 |
| 1 | 40 (46.0) | 54 (66.7) | Ref. | Ref. | ||
| 3 | 47 (54.0) | 27 (33.3) | 0.43 (0.23 - 0.80) | - | 0.36 (0.18 - 0.71) | - |
| rs1127354 | - | - | - | < 0.001 c | - | < 0.001 |
| CC | 57 (65.5) | 79 (97.5) | Ref. | - | Ref. | - |
| CA + AA | 30 (34.5) | 2 (2.5) | 0.05 (0.01 - 0.21) | - | 0.04 (0.01 - 0.19) | - |
| rs7270101 | - | - | - | 0.829 c | - | - |
| AA | 74 (85.1) | 70 (86.4) | Ref. | - | - | - |
| AC | 13 (14.9) | 11 (13.6) | 0.90 (0.37 - 2.13) | - | - | - |
| rs6051702 | - | - | - | 0.210 c | - | - |
| AA | 62 (71.3) | 65 (80.2) | Ref. | - | - | - |
| AC + CC | 25 (28.7) | 16 (19.8) | 0.61 (0.30 - 1.25) | - | - | - |
| RBV dose, mg/day | - | - | - | 0.361 d | - | - |
| 800 | 28 (32.9) | 22 (27.5) | - | - | - | - |
| 1000 | 28 (32.9) | 35 (43.8) | - | - | - | - |
| 1200 | 29 (34.1) | 23 (28.7) | - | - | - | - |
aValues are expressed as No. (%) unless otherwise indicated.
bMann-Whitney U-test.
cFisher-exact test.
dPearson chi-squared.
4.4. Evaluation of Factors Affecting Platelet -Decline at Week 4 of Treatment With Pegylated-Interferon Plus Ribavirin
In univariate analysis, no association was observed between the age, sex, BMI, ALT, AST, HCV genotype, rs7270101, rs6051702 and RBV dose with Plt-decline at week 4 of the treatment (P > 0.05) (Table 3). However, the HCV RNA level < 600,000 IU/mL (P = 0.010, OR = 3.45) and rs1127354 CA + AA (P = 0.002, OR = 4.22) were found to be associated with higher Plt-decline at week 4 of the treatment (Table 3). Multivariate analysis confirmed that both the HCV RNA level < 600,000 IU/mL (P = 0.012, OR = 3.45) and rs1127354 CA + AA (P = 0.002, OR = 4.89) were associated with greater Plt-decline at week 4 of the treatment (Table 3).
| Variables | Plt-Decline | Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|---|---|
| ≤ 8 × 104 (n = 142) | > 8 × 104 (n = 26) | OR (95% CI) | P Value | Adjusted OR (95% CI) | Adjusted P Value | |
| Age, median (IQR) | 40 (18.0) | 39 (17.0) | - | 0.374 b | - | - |
| Gender | 0.457 c | |||||
| Male | 131 (92.3) | 23 (88.5) | Ref. | - | - | |
| Female | 11 (7.7) | 3 (11.5) | 1.55 (0.40 - 6.00) | |||
| BMI, median (IQR) | 26.3 (5.6) | 27.0 (7.5) | - | 0.771 b | - | - |
| ALT, median (IQR), IU/L | 72 (76.0) | 66 (67.0) | - | 0.360 b | - | - |
| AST, median (IQR), IU/L | 41 (33.0) | 45 (31.0) | - | 0.811 b | - | - |
| HCV RNA level, IU/mL | 0.010 c | 0.012 | ||||
| > 600,000 | 92 (67.2) | 9 (37.5) | Ref. | Ref. | ||
| < 600,000 | 45 (32.8) | 15 (62.5) | 3.45 (1.38 - 8.33) | 3.45 (1.32 - 9.09) | ||
| HCV genotype | 0.138 c | 0.144 | ||||
| 1 | 83 (58.5) | 11 (42.3) | Ref. | Ref. | ||
| 3 | 59 (41.5) | 15 (57.7) | 1.92 (0.82 - 4.47) | 2.04 (0.78 - 5.32) | ||
| rs1127354 | 0.002 c | 0.002 | ||||
| CC | 121 (85.2) | 15 (57.7) | Ref. | Ref. | ||
| CA + AA | 21 (14.8) | 11 (42.3) | 4.22 (1.71 - 10.45) | 4.89 (1.79 - 13.37) | ||
| rs7270101 | 0.769 c | |||||
| AA | 122 (85.9) | 22 (84.6) | Ref. | - | - | |
| AC | 20 (14.1) | 4 (15.4) | 1.11 (0.35 - 3.56) | |||
| rs6051702 | 0.216 c | |||||
| AA | 110 (77.5) | 17 (65.4) | Ref. | - | - | |
| AC + CC | 32 (22.5) | 9 (34.6) | 1.82 (0.74 - 4.47) | |||
| RBV dose, mg/day | 0.407 d | |||||
| 800 | 40 (28.8) | 10 (38.5) | - | - | - | |
| 1,000 | 56 (40.3) | 7 (26.9) | ||||
| 1,200 | 43 (30.9) | 9 (34.6) | ||||
aValues are expressed as No. (%) unless otherwise indicated.
bMann-Whitney U-test.
cFisher-exact test.
dPearson chi-squared.
5. Discussion
Recently, studies showed that host genetic factors including ITPA SNPs and polymorphisms near IFNL3 play a great role in treatment of CHC (10, 12-22). Previous studies among Caucasian populations revealed that the minor allele frequency (MAF) of ITPA rs1127354 and rs7270101 polymorphisms have been 4% - 14% and 11% - 24%, respectively. In this study, we found that the MAFs of rs1127354 and rs7270101 were 10.1% and 7.1%, respectively, which are almost comparable to previous Caucasian-based reports (8, 10-12, 23, 24). Our study showed that the MAF for C20orf194 rs6051702 polymorphism was 14.6% and this finding is similar to the other Caucasian-based reports (14.5% - 19.2%) (10, 25). In the present study, we found that the LD between rs1127354 and rs7270101 with rs6051702 were lower than the other Caucasian populations (10).
To date, in our region the standard of care for CHC is the combination therapy with PEG-IFN plus RBV. The PEG-IFN acts as antiviral and immunomodulator and impacts both adaptive and innate immune responses against HCV. Ribavirin is a nucleoside analogue of guanosine, which interrupts the HCV RNA metabolism (4, 14). This combination therapy has a broad range of side-effects including gastrointestinal disturbances, influenza-like symptoms, fatigue, neuropsychiatric syndromes and hematological changes (26, 27). Previous studies showed that prescribing PEG-IFN plus RBV may cause cytopenia (4, 28). Hemoglobin-decline is a common side-effect of this regimen and it has been reported that more than 50% of the patients experience it and this issue leads the physicians for dose modification or termination of therapy (4, 15). Dose modification of RBV affect final outcome of treatment, which is the eradication of infection (4, 5, 27). Although previous studies showed that treatment-induced Hb-decline may be affected by age (29, 30), gender (29-31), baseline Hb (29), baseline Plt (32), plasma RBV concentration (33) and RBV dose (30), the most acceptable factors which influenced treatment-induced Hb-decline were rs1127354, rs7270101 and rs6051702 polymorphisms (10-12, 34).
The idea of ITPA polymorphisms play as a preventing factor for Hb-decline in the CHC patients during therapy with RBV-based regimens was firstly introduced by Fellay et al. (10) throughout the genome wide association study. They showed that several SNPs on chromosome 20 (20p13) were strongly related to treatment-induced Hb-decline. In this region of chromosome 20, the strongest signals were detected for ITPA rs1127354 and rs7270101 polymorphisms and also they found that these SNPs are associated with C20orf194 rs6051702 polymorphism (10). Subsequent studies confirmed this finding and revealed that minor alleles of rs1127354 and rs7270101 had the major role in preventing treatment-induced Hb-decline (34).
The rs1127354 and rs7270101 SNPs are functional ITPA gene polymorphisms, which subsequently concern a missense variant in exon 2 and a splicing-altering SNP in intron 2 of the ITPA gene. These two SNPs cause ITPase deficiency and subsequently result in accumulation of inosine triphosphate (ITP) in red blood cells (RBCs). On the other hand, RBV causes the depletion of adenosine triphosphate (ATP) which is followed by guanosine triphosphate deficiency in RBCs and causes RBC lysis. Accumulated ITP in ITPase-deficient patients would play a great role for compensating the depletion of ATP, and prevention of oxidative reactions and RBC lysis. This issue finally causes resistance potential for lower Hb-decline among ITPase-deficient patients in contrast to ITPase-sufficient patients (13, 35-38).
After performing univariate and multivariate analysis, we found that rs1127354 polymorphism was associated with Hb-decline at week 4 of treatment; however, no association was found between rs7270101 and rs6051702 and Hb-decline at week 4 of the combination therapy with PEG-IFN plus RBV. This finding is not in accordance with the results of most of the similar studies and it reveals that the most informative polymorphism among Iranian CHC patients for predicting Hb-decline at week 4 of treatment with PEG-IFN plus RBV is rs1127354. This finding may be caused by the population size, and it is suggested that further studies with larger sample size should be conducted. Another factor that we found to be effective on Hb-decline was the HCV genotype. Almost most of the studies in this field showed no significant association between Hb-decline and HCV genotypes. This issue may be caused by homogeneity of the HCV genotype in previous studies (16, 39, 40). Maan et al. (41) showed that although in univariate analysis there was a significant association between the HCV genotype (-1/4 versus -2/3) and Hb-decline at week 4 of treatment with PEG-IFN plus RBV, in multivariate analysis they found no significant association between the HCV genotypes and Hb-decline (41). In the current study, we found that patients with the HCV genotype-1 had a higher rate of Hb-decline in contrast to patients with the HCV genotype-3. The idea of different prescribing dose of RBV among HCV genotype-1 and -3 led us to assess this issue. After we found there was no significant association between the RBV dose and Hb-decline, we suggested that the HCV genotype may be an independent factor influencing Hb-decline at week 4 of treatment with PEG-IFN plus RBV.
Some studies showed that although ITPase-deficient patients protected against RBV-induced Hb-decline, these patients experienced higher Plt-decline in contrast to patients with normal ITPase activity (8, 9). It has been suggested that patients with normal ITPase activity have a greater chance to compensate thrombocytopenia in reaction to higher Hb-decline (8, 42-46). Current study revealed that Plt-decline may be affected by rs1127354 CA + AA, the condition which is strongly related with preventing Hb-decline at week 4 of treatment with PEG-IFN plus RBV. We suggest that lower rate of Plt-decline among patients with rs1127354 CC which had the higher rate of Hb-decline, may be caused by reactive response which compensates thrombocytopenia event. Furthermore, another factor that seemed to have effect on Plt-decline in our study was the HCV RNA level. Patients with HCV RNA < 600,000 IU/mL had a higher rate of Plt-decline in contrast to patients with HCV RNA > 600,000 IU/mL. This finding may be caused by the sample size and it can be suggested that further studies with larger sample size should be conducted.
New treatments for hepatitis C have been introduced in recent years. These treatment regimens have higher efficacy and lower side-effects; however, they are not affordable in developing countries because of their high-cost. It is worth to say almost the frequency of HCV infection is higher in the developing countries, cost limitation leads remaining of PEG-IFN plus RBV as the most prescribing regimen in these countries.
In conclusion, in addition to the assessment of routine biomarkers prior to treatment with PEG-IFN plus RBV, genotyping of ITPA rs1127354 polymorphism would be beneficial for predicting the Hb-decline among patients with CHC. Furthermore, ITPA rs1127354 polymorphism and also the baseline HCV RNA level could be defined as predictors for Plt-decline during treatment with PEG-IFN plus RBV in patients with CHC.