1. Background
Chronic hepatitis C (CHC) is a worldwide infection caused by the hepatitis C virus (HCV), an enveloped RNA virus. It affects more than 170 million people all over the world (1). After an often asymptomatic acute infection, approximately 70% of all infected patients progress to chronic hepatitis, which often results in long-term and permanent sequelae; HCV infection is therefore one of the most common causes of chronic liver disease and hepatocellular carcinoma (HCC) (2). The prevalence of HCV infection varies worldwide. It has been reported to range from 0.2% to 2.2% in developed countries and is less than 0.5% in Iran (0.1% in women and 1% in men) (3-9).
HCV is categorized into 7 distinct genotypes (1-7) and 67 subtypes (e.g., 1a, 3a) based upon differences in its genome (4, 6, 10, 11). The prevalence of these HCV genotypes varies in different geographic areas. HCV subtypes 1a and 3a are the most common subtypes in Iran (4, 6-8, 11-16). Among Iranian hemophilic and thalassemic patients, HCV subtype 1a is also the most prevalent, followed by subtypes 3a and 1b (17, 18). The HCV genome encodes a large poly-protein, which is finally processed to structural proteins, such as the core protein and non-structural proteins, including NS3 protease (19-21). The core protein is a highly conserved basic protein with 191 amino acids (aa), which forms a viral nucleocapsid, interacts with numerous cellular proteins, and affects several host cell functions (e.g., gene transcription, apoptosis, and signaling pathways) (4, 6, 10, 11).
Recent studies have suggested several new prognostic molecular factors in hepatitis C patients, which could be categorized into viral and host factors (2, 19-22). Viral factors include the HCV genotype and aa substitution in HCV proteins. One of the most studied variations in HCV is the core aa 70 substitution. HCV subtypes 3a and 1b are associated with cirrhosis and HCC, respectively; aa substitution of arginine by glutamine at the core 70 position (Arg70Gln) has been correlated with a poor therapeutic response, HCC, and liver-related death (2, 19-22). Host factors include gender, age, and polymorphisms near the IFNL3 gene, such as rs12979860 and rs8099917. Male gender and an age > 60 have been identified as determinants of hepatocarcinogenesis and impact on the survival rates for liver-related death (22).
Although new therapeutic regimens with great efficacy have been developed for CHC management, the high costs of these regimens limit their widespread application. Nevertheless, in many countries, interferon-based regimens have remained the standard of care. Many previous studies have shown that the rs12979860 CC and rs8099917 TT genotypes are associated with a higher probability of spontaneous clearance and therapeutic response (2, 19-26). Previous studies have also suggested the effects of these factors on other baseline factors (27).
Hepatitis C is a transfusion-transmitted infection; therefore, multitransfused patients (e.g., thalassemic and hemophilic patients) are at a higher risk of infection (28-32). The prevalence of HCV infection and HCV genotypes among blood product recipients and individuals who take part in high-risk behaviors, like intravenous drug use (IDU) and tattooing have been studied in several countries and revealed significantly higher rates of HCV infection among these susceptible groups. Although HCV-infected hemophilic and thalassemic patients have been examined regarding HCV infection and genotype prevalence, their molecular characteristics (including the core aa 70 substitution and polymorphisms near the IFNL3 gene) have not been clearly identified.
2. Objectives
This study aimed to determine the molecular profile of HCV infection (including the HCV genotype, core aa 70 substitution, and rs12979860 and rs8099917 polymorphisms near the IFNL3 gene) among Iranian patients with CHC infection in order to evaluate the impacts of these factors on disease outcome and the necessity of commencing appropriate treatment.
3. Patients and Methods
3.1. Study Population
We recruited 141 hemophilic, 84 thalassemic, and 204 non-hemophilic, non-thalassemic (NHNT) patients who were chronically infected with HCV. The NHNT patients had hepatitis C risk factors, such as IDU, a past history of blood transfusion and prior tattooing. These patients had been referred to either the Tehran blood transfusion hepatitis clinic (affiliated with the Iranian Blood Transfusion Organization (IBTO)) or the Tehran hepatitis clinic (affiliated with the Baqiyatallah Research Center for Gastroenterology and Liver diseases (BRCGL)) between December 2009 and November 2015. The study was approved by the local ethics committee of Baqiyatallah University of Medical Sciences. Inclusion criteria were defined as a positive result for anti-HCV antibodies along with a positive result for plasma HCV RNA by reverse transcription-polymerase chain reaction (RT-PCR) for more than six months. Exclusion criteria included hepatitis B virus or human immunodeficiency virus co-infection. All patients were Iranian and of Caucasian ethnicity.
3.2. Laboratory Assessment
Assessment of the HCV RNA level was carried out using COBAS® TaqMan® HCV Test v2.0 (Roche Diagnostics) according to the manufacturer’s instructions. Nested RT-PCR followed by DNA sequencing were used for the assessment of HCV core aa 70 substitutions. Briefly, viral RNA was extracted using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The extracted viral RNA was subjected to cDNA synthesis using a RevertAid First Strand cDNA Synthesis Kit (Fermentas of Thermo Fisher Scientific, Waltham, MA, US). The nested RT-PCR was performed using an Accupower PCR PreMix (Bioneer Corp., Daejeon, South Korea) with the following conditions for both the first and nested rounds of PCR: 94°C for 5 minutes, 35 cycles at 94°C for 30 seconds, 50°C for 40 seconds, and 72°C for 40 seconds, followed by 72°C for 5 minutes. The primers were adopted from the study by Kumar et al. (33). Primers used for the first round reaction included the following: HG1, 5’-ATGTACCCCATGAKRTCGGC-3’ and HG2, 5’-ACTGCCTGATAGGGTGCTTGCGAG-3’; the nested round reaction used the following primers: HG3, 5’-CAYGTRAGGGTATCGATGAC-3’ and HG4, 5’-AGGTCTCGTAGACCGTGCATCATG-3’. The 404 base pair (bp) amplified products of the nested RT-PCR were purified using a QIAquick PCR Purification Kit (Qiagen, Hilden, Germany), followed by DNA sequencing with a Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, US) and a 3130XL ABI Genetic Analyzer (Applied Biosystems, Foster City, CA, US). The sequence traces were assessed using a CLC Sequence Viewer (version 6), and the DNA sequences were translated to protein to detect any changes at the 70 position of the HCV core protein. The HCV genotyping was performed based on phylogenetic analysis using MEGA 5 software and the neighbor-joining tree and Kimura 2-parameter models (The phylogenetic tree will not be represented in the results section of this study). The rs12979860 and rs8099917 polymorphisms were assessed as the most common polymorphisms near the IFNL3 gene. The detailed protocol of the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method for genotyping the rs12979860 and rs8099917 polymorphisms was as previously described (34). The latest laboratory data of the serum alanine aminotransferase (ALT) level (IU/L) was recruited from patient records.
3.3. Liver Fibrosis Assessments
Patients were assessed regarding the development of cirrhosis via liver biopsy or alternative methods. In patients who underwent a liver biopsy, liver specimens were processed using standard criteria. Staging of fibrosis was determined using a modified Knodell scoring system (35). A liver biopsy stage ≥ 5 was considered to indicate severe liver fibrosis (cirrhosis). The liver histology was evaluated by an expert pathologist who was blinded to the study’s design. Not all patients underwent a liver biopsy because it is not necessary to determine the histological severity before initiating HCV treatment; in addition, this procedure can be life threatening and is costly in patients with congenital bleeding disorders. Liver stiffness was assessed by transient elastography using a FibroScan 502 machine (EchoSense) for some of the patients, and the results were defined as F0 - F4; any score > F3 or > 12.5 Kpa was considered severe fibrosis or cirrhosis. For instances where a FibroScan was not accessible, evidence of liver cirrhosis was defined based on the available clinical and imaging evidence.
3.4. Statistical Analysis
Data analysis was performed using the Statistical Package for the Social Sciences (SPSS) (version 16). The odds ratio (OR) (with 95% confidence intervals) and P values were calculated for each allele and genotype and were tested using a χ2 test. Fisher’s exact or Monte Carlo tests were performed for χ2 tests when more than 20% of the cells showed less than the expected frequency of five. A haplotype analysis was carried out with SHEsis software (36). The distributions of genotypes of polymorphisms near the IFNL3 gene were tested for deviations from Hardy-Weinberg equilibrium (HWE). For continuous variables, a one-way analysis of variance (ANOVA) or Kruskal-Wallis, and independent t-test or Mann-Whitney U-test were used to compare groups according to the results of the Shapiro-Wilk test. P values less than 0.05 were considered statistically significant. The Graphpad Prism (version 6) software was applied to draw graphs and representations.
4. Results
4.1. Patient Characteristics
Table 1 shows the differences in the demographics and clinical characteristics of the studied patients. Although cirrhosis was observed more frequently in patients with HCV subtype 1b (25%) than in those with HCV subtypes 1a (20.6%) and 3a (17.2%), no statistically significant difference was found in respect to the frequency of cirrhosis between HCV subtypes regardless of their previous treatment history (P = 0.796). Within the NHNT patient group, IDU was observed in 107 patients and was significantly associated with HCV genotypes (P = 0.032); therefore, HCV subtype 3a was significantly more frequent in IDU patients (43.0%) compared to non-IDU (27.1%) ones.
| Characteristics | Patient Category | P Value | |||
|---|---|---|---|---|---|
| Total (n = 429) | Hemophilia (n = 141) | Thalassemia (n = 84) | Non-Hemophilia, Non-Thalassemia (n = 204) | ||
| Age, ya | 33 (14) | 31 (14) | 27 (7) | 39 (18) | < 0.001b |
| Gender (male), No. (%) | 368 (85.8) | 131 (92.9) | 52 (61.9) | 185 (90.7) | < 0.001c |
| Treatment history, No. (%) | 175 (40.8) | 70 (49.6) | 50 (59.5) | 55 (28.4) | < 0.001c |
| HCV RNA level, log IU/mLa | 6.10 (0.99) | 6.17 (0.82) | 5.75 (1.08) | 6.11 (1.05) | < 0.001b |
| HCV subtypes, No. (%) | |||||
| 1a | 254 (59.2) | 94 (66.7)d | 50 (59.5) | 110 (53.9) | 0.018c,d |
| 1b | 44 (10.3) | 21 (14.9) | 4 (4.8)d | 19 (9.3) | 0.026c,d |
| 3a | 123 (28.7) | 23 (16.3)d | 28 (33.3) | 72 (35.3) | < 0.001c,d |
| Cirrhosis, No. (%) | 74 (17.2) | 17 (13.2) | 34 (43.6) | 23 (14.5) | < 0.001c |
| Core aa 70 substitution, No. (%) | 0.690e | ||||
| Total | 44 (10.3) | 17 (12.1) | 8 (9.5) | 19 (9.3) | |
| Arg70Gln | 36 (8.4) | 14 (9.9) | 8 (9.5) | 14 (6.9) | |
| Arg70His | 3 (0.7) | 0 (0) | 0 (0) | 3 (1.5) | |
| Arg70Pro | 4 (0.9) | 2 (1.4) | 0 (0) | 2 (1) | |
| Arg70Leu | 1 (0.2) | 1 (0.7) | 0 (0) | 0 (0) | |
| rs12979860, No. (%) | |||||
| Genotype | 0.060c | ||||
| CC | 141 (33.1) | 39 (27.9) | 22 (26.2) | 80 (39.6) | |
| CT | 210 (49.3) | 75 (53.6) | 42 (50.0) | 93 (46.0) | |
| TT | 75 (17.6) | 26 (18.6) | 20 (23.8) | 29 (14.4) | |
| Allele | 0.019c | ||||
| C | 492 (57.7) | 153 (54.7) | 86 (51.2) | 253 (62.5) | |
| T | 360 (42.3) | 127 (45.3) | 82 (48.8) | 151 (37.5) | |
| rs8099917, No. (%)f | |||||
| Genotype | 0.268c | ||||
| TT | 201 (53.9) | 66 (50.8) | 36 (46.8) | 99 (59.6) | |
| GT | 146 (39.1) | 55 (42.3) | 33 (42.9) | 58 (34.9) | |
| GG | 26 (7.0) | 9 (6.9) | 8 (10.4) | 9 (5.4) | |
| Allele | 0.092c | ||||
| T | 548 (73.5) | 187 (71.9) | 105 (68.3) | 256 (77.1) | |
| G | 198 (26.5) | 73 (28.1) | 49 (31.7) | 76 (22.9) | |
aData are expressed as median (interquartile range) due to deviation from the normal distribution.
bKruskal-Wallis test.
cχ2 test.
dPost hoc test.
eFisher’s exact test.
fMissing data were less than 10% for all variables, except for rs8099917, which was 14.5%.
The core aa 70 substitution of HCV was not significantly different across patient groups (Table 1). However, hemophilic patients showed the highest rate of substitution (12.1%). Moreover, Arg70Gln was significantly associated with an increased risk of cirrhosis (OR = 2.49; 95% CI = 1.13 - 5.50; P = 0.020).
Concerning the frequencies of genotypes and alleles of polymorphisms near IFNL3 (rs12979860 and rs8099917), no significant deviation from HWE was observed (rs12979860, P = 0.87; rs8099917, P = 0.89). More interestingly, the minor T allele of rs12979860 showed significantly increased frequencies in the hemophilia and thalassemia groups, as compared with the NHNT patient group (hemophilia vs. NHNT: OR = 1.39, 95% CI = 1.02 - 1.89, P = 0.036; thalassemia vs. NHNT: OR = 1.59, 95% CI = 1.11 - 2.29, P = 0.011). However, a significantly increased frequency for the rs8099917 G allele was only observed in thalassemic patients when compared to NHNT patients (OR = 1.57, 95% CI = 1.03 - 2.40, P = 0.036). Furthermore, thalassemic patients showed significantly different frequencies of haplotypes as compared to NHNT patients (CT haplotype OR = 0.61, 95% CI = 0.41 - 0.89, P = 0.011; TG haplotype OR = 1.60, 95% CI = 1.05 - 2.46, P = 0.028). The linkage disequilibrium between rs12979860 and rs8099917 was calculated, and D’ was 1.0 with r2 = 0.476.
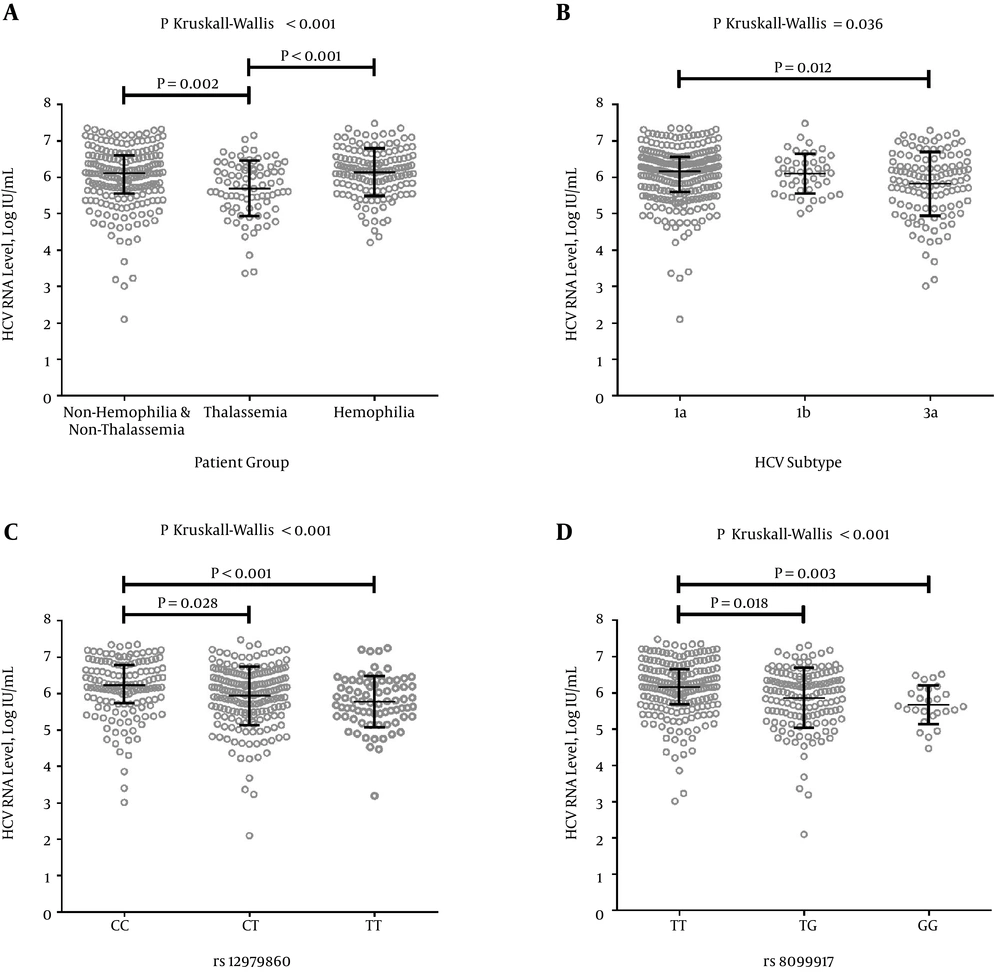
Regarding the clinical impact of molecular variations of HCV infection, significantly lower HCV RNA levels were found in patients infected with HCV subtype 3a than HCV subtypes 1a and 1b (Figure 1B). Furthermore, the patients with rs12979860 TT and rs8099917 GG genotypes showed significantly lower HCV RNA levels than other rs12979860 and rs8099917 genotypes, respectively (P < 0.001; Figure 1C and 1D). The level of ALT in patients with the rs12979860 CC genotype was significantly higher than in patients with the other rs12979860 genotypes (P = 0.026). Moreover, the ALT levels of patients with HCV subtype 3a was the highest, followed by HCV subtypes 1a and 1b (P < 0.001).
Association of HCV RNA level and characteristics of HCV-infected patients. A) Differences in log-transformed HCV RNA levels between patient groups; B) Differences in log-transformed HCV RNA levels among common HCV subtypes; C) Differences in log-transformed HCV RNA levels among genotypes of rs12979860; and D) Differences in log-transformed HCV RNA levels among genotypes of rs8099917. Post hoc tests were conducted for assessments between groups. Median and interquartile range (IQR) are represented with line and bar, respectively, for each group.
4.2. Association of HCV Genotype With Core aa 70 Substitution
Among all 429 HCV-infected patients, the HCV subtype was found to be significantly associated with a core aa 70 substitution of HCV (P < 0.001) so that Arg70Gln was observed more frequently in HCV subtype 1b (54.5%) compared to HCV subtypes 1a (1.2%) and 3a (4.9%). Furthermore, a significantly strong correlation between HCV subtype 1b and core aa 70 substitution was revealed (Kendall’s tau = 0.532, P < 0.001). Table 2 demonstrates the frequencies of core aa 70 substitution in HCV subtypes.
| Subtypes | Core aa 70 Substitution | Total | ||||
|---|---|---|---|---|---|---|
| Arginine | Glutamine | Histidine | Proline | Leucine | ||
| HCV, No. (%) | ||||||
| 1a | 246 (96.8) | 3 (1.2) | 0 (0) | 4 (1.6) | 1 (0.4) | 254 |
| 1b | 18 (40.9) | 24 (54.5) | 2 (4.5) | 0 (0) | 0 (0) | 44 |
| 3a | 116 (94.3) | 6 (4.9) | 1 (0.8) | 0 (0) | 0 (0) | 123 |
| 3k | 2 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 |
| 4 | 3 (50.0) | 3 (50.0) | 0 (0) | 0 (0) | 0 (0) | 6 |
| Total | 385 | 36 | 3 | 4 | 1 | 429 |
aP < 0.001 (with Fisher’s exact test).
4.3. Core aa 70 Substitution and rs12979860 and rs8099917 Polymorphisms
The genotype frequencies of polymorphisms near IFNL3 were significantly different regarding core aa 70 substitution (rs12979860, P = 0.045; rs8099917, P = 0.024) (Table 3), so that 33.3% of patients with glutamine at the HCV core aa 70 had the rs12979860 TT genotype, as compared to 16.2% of patients with arginine at this position.
| Genotypes | Core aa 70 | P Value | |||||
|---|---|---|---|---|---|---|---|
| Arginine | Glutamine | Histidine | Proline | Leucine | Total | ||
| rs12979860, No. (%) | 0.045a | ||||||
| CC | 130 (33.9) | 7 (19.4) | 1 (50.0) | 2 (50.0) | 1 (100) | 141 | |
| CT | 191 (49.9) | 17 (47.3) | 0 (0) | 2 (50.0) | 0 (0) | 210 | |
| TT | 62 (16.2) | 12 (33.3) | 1 (50.0) | 0 (0) | 0 (0) | 75 | |
| Total | 383 | 36 | 2 | 4 | 1 | 426 | |
| rs8099917, No. (%) | 0.024a | ||||||
| TT | 187 (55.8) | 12 (36.3) | 0 (0) | 1 (33.3) | 1 (100) | 201 | |
| GT | 128 (38.2) | 16 (48.5) | 0 (0) | 2 (66.7) | 0 (0) | 146 | |
| GG | 20 (6.0) | 5 (15.2) | 1 (100) | 0 (0) | 0 (0) | 26 | |
| Total | 335 | 33 | 1 | 3 | 1 | 373 | |
aAccording to Fisher’s exact test.
Furthermore, it was observed that a mutant core aa 70 (amino acids other than arginine) was more common among rs8099917 non-TT genotypes versus the TT genotype (14.0% vs. 7.0%, P = 0.026); a negative mild correlation was also found between the rs8099917 TT genotype and the mutant core aa 70 (Kendall’s tau = -0.115, P = 0.038). The same was observed for the rs12979860 TT genotype vs. the non-TT genotypes (17.3% vs. 8.5%, P = 0.022) and produced a mild correlation between the rs12979860 TT genotype and the mutant core aa 70 (Kendall’s tau = 0.111, P = 0.032).
4.4. Correlation Between HCV Genotypes and Polymorphisms Near the IFNL3 Gene
Different frequencies of the rs8099917 genotypes were found in respect to the HCV genotype (P = 0.021) so that the rs8099917 GG genotype was more common in the HCV subtype 1b-infected patients group (13.2%) in comparison with HCV subtypes 1a (6.4%) and 3a (5.5%). Different frequencies were also observed for the rs12979860 CC genotype versus non-CC genotypes regarding HCV subtype (P = 0.036; Table 4) so that the rs12979860 CC genotype was more common in HCV subtype 3a-infected patients (43.1%) compared to HCV subtypes 1a (30.2%) and 1b (23.2%). Additionally, significant mild negative correlations were found between the HCV subtype and polymorphisms near the IFNL3 gene (rs12979860, Kendall’s tau = -0.113, P = 0.001; rs8099917, Kendall’s tau = -0.105, P = 0.006).
| Genotypes | HCV Subtypes | Total | P Value | ||||
|---|---|---|---|---|---|---|---|
| 1a | 1b | 3a | 3k | 4 | |||
| rs12979860, No. (%) | 0.097a | ||||||
| CC | 76 (30.2) | 10 (23.2) | 53 (43.1) | 1 (50.0) | 1 (16.7) | 141 | |
| CT | 134 (53.1) | 22 (51.2) | 50 (40.6) | 1 (50.0) | 3 (50.0) | 210 | |
| TT | 42 (16.7) | 11 (25.6) | 20 (16.3) | 0 (0) | 2 (33.3) | 75 | |
| Non-CC, No. (%) | 176 (69.8) | 33 (76.8) | 70 (56.9) | 1 (50.0) | 5 (83.3) | 285 | 0.036a,b |
| rs8099917, No. (%) | 0.021a | ||||||
| TT | 112 (51.4) | 17 (44.7) | 71 (64.5) | 1 (50.0) | 0 (0) | 201 | |
| GT | 92 (42.2) | 16 (42.1) | 33 (30.0) | 1 (50.0) | 4 (80.0) | 146 | |
| GG | 14 (6.4) | 5 (13.2) | 6 (5.5) | 0 (0) | 1 (20.0) | 26 | |
| Non-TT, No. (%) | 106 (48.6) | 21 (55.3) | 39 (35.5) | 1 (50.0) | 5 (100) | 172 | 0.006a,c |
aAccording to Fisher’s exact test.
bIn comparison to the rs12979860 CC genotype.
cIn comparison to the rs8099917 TT genotype.
5. Discussion
The main findings of this study were as follows: 1) Arg70Gln was the most common substitution of core aa 70, which was significantly associated with an increased risk of cirrhosis; 2) Arg70Gln was strongly correlated with the HCV subtype 1b; 3) Core aa 70 substitutions were more common in the rs12979860 TT genotype and the rs8099917 non-TT genotypes, and a mild correlation was found between core aa 70 substitution and polymorphisms near the IFNL3 gene; 4) Lower HCV RNA levels were significantly associated with the rs12979860 TT and the rs8099917 GG genotypes as well as HCV subtype 3a; 5) the rs8099917 GG and rs12979860 CC genotypes were observed more frequently in patients infected with HCV subtypes 1b and 3a, respectively.
To the best of our knowledge, HCV subtype 1a has been previously demonstrated as the most common in Iran (4, 6, 12, 37), and this prevalence was confirmed in this study. Few studies (Isfahan province (14) and Yazd province (38), central Iran; and Mashhad, northeast Iran (7)) have reported HCV subtype 3a as the most common (61.2%, 50.3%, and 40%, respectively). The different age of the studied patients could justify these differences, as it has been shown that HCV subtype 3a is more common in patients younger than 40 years (41.5%) (12). Studies from other countries have also confirmed an association between a young age and HCV subtype 3a (39-42). One study on Iranian HCV-infected thalassemic patients in central Iran also revealed that HCV subtype 1a is the most common genotype (15). On the other hand, a significantly higher frequency of HCV subtype 1b was identified in hemophilic patients, which was compatible with the findings of a previous study (18).
The results of the present study regarding core aa 70 substitutions in Iranian HCV-infected patients demonstrated an association of Arg70Gln with an increased risk of cirrhosis; interestingly, previous evidence supports the observations of this study (43). Based on these findings, Arg70Gln could be implied as a plausible predictor for cirrhosis and severe liver disease, especially when associated with HCV subtype 1b (44). It is worthwhile to note that from a molecular perspective, a probable role has been suggested for core protein in HCV-related cirrhosis via the up-regulation of vascular endothelial growth factor expression (45, 46). Interestingly, some rare substitutions at core aa 70 have been observed; 18% of core aa 70 substitutions included histidine, proline, or leucine.
In this study, it was revealed that Arg70Gln was significantly associated with HCV subtype 1b. While most of the previous studies on the therapeutic impacts of HCV subtype 1b have been conducted in Japan, a few other investigations have also reported high frequencies of Arg70Gln in patients infected with HCV subtype 1b in different populations, such as in Venezuela, the United States, and China (47). The Arg70Gln frequency in HCV subtype 1b was reported within a wide range, from 10% in China to 79% in Venezuela, and is compatible with the reported frequency in this study (54.5%) (47). Jaspe et al. (47) did not report any frequencies of Arg70Gln in HCV subtypes 1a and 3a that were higher than 3.7% and 11%, respectively, which were similar to the findings of the present study. An association of Arg70Gln with HCV subtype 1b has been reported along with a higher risk of poor treatment response (44, 48), lipid accumulation and steatosis, and an increased risk of HCC and insulin resistance (46). Although a higher rate of HCC is reported in patients with eradicated HCV subtype 1b infection associated with core Arg70Gln (49), early therapeutic intervention with an appropriate regimen to confront the infection by HCV subtype 1b associated with core Arg70Gln may improve the outcome and prevent long-term sequelae. Future studies are required to clarify these impacts. The high frequency of core aa 70 substitution that was found in hemophilic patients could also be due to the increased frequency of HCV subtype 1b within hemophilic patients and the association between HCV subtype 1b and core aa 70 substitution (47).
The genetic polymorphisms located near the IFNL3 gene and core aa 70 substitutions of HCV have been demonstrated to be predictive of the treatment response and prognosis (2, 19, 20, 25, 50). Based on a recent meta-analysis, the rs12979860 CC and rs8099917 TT genotypes have been associated with a higher probability of sustained virological response (SVR) (51). These genotypes have been also correlated with higher HCV RNA levels in the absence of treatment (52-54). Along the same lines, the rs12979860 TT and rs8099917 GG genotypes were correlated with lower HCV RNA levels in this study of HCV-infected patients. Lower HCV RNA levels in patients with the rs12979860 TT genotype were also previously reported by Haj-Sheykholeslami et al. (25). This finding could be interpreted in light of the higher expression of interferon alpha and beta genes and also interferon-stimulated genes in patients with the rs12979860 TT genotype (55-57).
Concerning the genotype frequencies of polymorphisms near the IFNL3 gene in Iranian patients, the rs12979860 CC, CT, and TT genotype frequencies were reported to be as high as 38%, 48.8%, and 13.2%, respectively, by Sharafi et al. (24). Additionally, the frequencies of the rs8099917 TT, GT, and GG genotypes were 58.3%, 37.1%, and 4.6%, respectively (24). The frequencies of these aforementioned genotypes observed in this study were similar to those in the mentioned report (24).
It is of particular importance to note that the thalassemic patients in this study showed a higher frequency of the rs12979860 T allele, which has been demonstrated to be associated with a poor treatment response (2, 19). In addition, the lower HCV RNA level found in thalassemic patients might be interpreted in the context of the higher prevalence of cirrhosis and also the probable role of hepatic iron deposition in hepatocyte metabolism, as well as an additional possible role for the core protein in respect to enhancing hepatocyte iron levels (58). Further prospective studies on separate naïve and non-naïve subgroups of HCV-infected thalassemic patients will be required to clarify the impacts of rs12979860 polymorphism on the course of CHC in thalassemic patients.
A significant link between core aa 70 substitution and the rs8099917 genotypes has been shown in previous studies, and treatment-resistant substitutions of core aa 70 (glutamine/histidine) were significantly more frequent in patients with treatment-resistant rs8099917 non-TT genotypes than the TT genotype (45, 59, 60). It has also been demonstrated that the cumulative change rates from arginine to glutamine/histidine in core aa 70 were significantly higher in rs8099917 non-TT genotypes than in TT genotypes (22). The present study agrees with these findings, as the rs8099917 non-TT genotypes had a higher frequency of core aa 70 substitutions; only a mild correlation was found. Interestingly, another correlation was also identified between the rs12979860 TT genotype and core aa 70 substitution. According to previous studies, the rs12979860 C allele is associated with a higher treatment response (54, 61-63). The association of these unfavorable factors may suggest an interaction between core aa 70 substitution and polymorphisms near the IFNL3 gene as well as a possible role in determining the disease outcome. Future studies are required to determine the existence and clarification of this interaction.
The previous study of Hajia et al. (11) on Iranian HCV-positive patients showed that HCV subtype 3a had a 30.76% frequency of high HCV RNA levels (≥ 800,000 IU/mL), while 69.22% of patients infected with HCV genotype 1 had elevated HCV RNA levels. Additionally, Chakravarti et al. (64) reported that HCV genotype 1 had the highest HCV RNA level in comparison to HCV genotypes 2 and 3 in another Asian population in India. These findings agree with the present study in that HCV subtype 3a showed the lowest HCV RNA levels.
Based on recent meta-analyses, rs8099917 TT genotype and T allele contribute to higher rates of SVR (65) and spontaneous HCV clearance in Caucasians mainly in individuals infected with HCV genotype 1 (66). This study demonstrated an association between rs8099917 GG genotype and HCV subtype 1b, which is in line with previously mentioned studies and could be due to the aforementioned contributory role of the rs8099917 TT genotype and T allele. The finding of a higher frequency of the rs12979860 CC genotype in HCV subtype 3a-infected patients of this study was also in agreement with the results of a recent meta-analysis study, which reported an association of the rs12979860 CC genotype and C allele with HCV subtype 3a (67), and also with similar findings of other studies on Iranian populations (24, 26).
We are witnessing a rapid evolution in the treatment of CHC (68). Emerging direct-acting antivirals (DAAs) have been taking promising steps towards improving the efficacy of therapeutic regimens, augmenting SVR rates to ≥ 90% (69), and countervailing the low success rates and severe adverse reactions of previous treatments (70). Selecting an appropriate therapeutic regimen based on the viral and host characteristics is emerging as the best practice routine for the management of HCV infection, but further studies and randomized clinical trials are still required in Iranian patients.
In this study, we faced some limitations; it was not possible to design a prospective study to assess the links between the molecular characteristics of HCV infection and the disease prognosis. Another limitation was the inevitable impact of possible time-related changes in HCV molecular characteristics, such as the substitution of core aa 70 after current assessment in this study, as the study population included both naïve and treatment-experienced patients and confronted the authors with a significant difference between these two groups regarding core aa 70 substitution and the rs12979860 genotypes, especially in hemophilic patients.
In conclusion, several associations between polymorphisms near the IFNL3 gene, HCV core aa 70 substitution and the HCV genotype were demonstrated. Moreover, it was found that HCV subtype 1a was the most prevalent subtype in Iranian patients with CHC. Arg70Gln was revealed to be the most common substitution and was significantly associated with an increased risk of cirrhosis. Additionally, the host characteristics (rs12979860 TT and rs8099917 GG genotypes) were linked with lower HCV RNA levels. Future studies seem reasonable, important, and essential to clarify any unrecognized and possibly important interactions between polymorphisms near the IFNL3 gene, the HCV genotype and core aa 70 substitution to reveal the future route of HCV infection therapy.