1. Background
Hepatitis C virus (HCV) is an enveloped, single-stranded RNA virus belonging to the Flaviviridae family (1, 2). It is estimated that more than 3% of the world’s population is infected with HCV (1). In many patients, chronic infection with HCV may cause liver disorders, such as cirrhosis, or more severe diseases, such as hepatocellular carcinoma (3). HCV is classified into seven major genotypes with more than 80 subtypes (4). According to various climatic conditions and transmission routes, different statistics are reported for the HCV genotype distribution in each country or region. In Iran, HCV genotypes 1a and 3a have been reported to be more frequent, with 41.7% and 33.0% prevalence rates, respectively (2). The HCV genome is a circular RNA that encodes a large, 3,000-amino-acid polyprotein that is cleaved into different structural and non-structural proteins (5). The four structural proteins are the core protein (22 kDa), the glycoproteins E1 (33 kDa) and E2 (70 kDa), and p7. The non-structural (NS) proteins are NS2, NS3, NS4A, NS4B, NS5A, and NS5B (6). Among the structural proteins encoded by HCV, the E2 glycoprotein plays an important role in the binding and entry of the virus into hepatic cells. As such, it has been shown that the production of neutralizing antibodies (NAb) against the E2 protein receptor binding sites prevents HCV infection in chimpanzees (7); this has effectively eliminated the virus in humans as well (8). Therefore, the induction of NAb against HCV E2 would be an effective strategy for the production of vaccines against the virus. The HCV E2 glycoprotein itself consists of three variable surface regions, including a highly variable region, the central region of the E2 protein, which is separated by disulfide bonds, and the intergenotypic variable region (igVR) which is located at the third region of the E2 protein, which is separated by disulfide bonds, and the intergenotypic variable region (igVR) are located at the third region of the E2 glycoprotein (amino acids 431 - 466) (9). The igVR is known to be involved in viral attachment to specific liver surface receptors (such as CD81 and SR-BI) and virus entry into the hepatic cells (10). Although the igVR is relatively the same among similar genotypes of HCV, it shows a high degree of variability between different genotypes (10). Currently, among the HCV genotypes, HCV genotype 2a (known as JFH1) has the ability to be cultured in vitro. Accordingly, there have been many efforts to create HCV chimeras and identify the mutations; however, the production of an attenuated vaccine against HCV is the main goal. One of the innovative aspects of this project was the construction of a combined virus of the two genotypes, using genetic engineering. The chimeric virus will be used in future studies for reverse genetic analysis to investigate the behavior of HCV in cell cultures.
The innovation of this project was to produce a chimera virus with a combination of genotypes 1a and 2a. The 1a genotype has a high prevalence in Iran and the 2a (JFH1) genotype has the ability to be cultured in vitro. Our study’s plan is in line with other large-scale projects to develop a vaccine for hepatitis C. This study was designed to take steps toward the production of a vaccine against this native virus in Iran (genotype 1). Evaluation of the immunogenicity of E2 glycoprotein peptides was another goal in this study. This region is important because it is the virus attachment site on the cell surface and affects gene sequence changes among genotypes.
2. Objectives
In the present study, we replaced the genotype 1 E2 variable glycoprotein regions with the same part in genotype 2.
3. Methods
3.1. Primer Design and PCR Amplification
In this study, we used plasmids pFL-J6/JFH (genotype 2a) and pSGR-JFH1 (GND). The pSGR-JFH1 (GND) plasmid has a point mutation in NS5B (G8624A), which can be used as a negative control. We also retrieved the genotype 1 sequence from the NCBI database and designed the primers based on this data. These plasmids were transferred into Escherichia coli strain JM109 and purified using a QIA prep Miniprep kit (Qiagen, USA) according to the manufacturer’s recommendations. The concentrations of the pFL-J6/JF and pSGR-JFH1 plasmids DNA were detected using the optical density (OD) method, as well as agarose gel electrophoresis.
3.2. Construction of the igVR-1a/JFH1 Chimera Genome
Two pairs of specific primers were designed for amplifying the chimeric region. These primers contained the BsiWI (Bold letters in the forward 1 primer) and KpnI (Bold letters in the reverse 2 primer) restriction enzyme cleavage sites (Table 1). Gene amplification was carried out using a thermal cycler system (Applied Biosystems, USA). All PCR products were checked using 1.5% agarose gel and were stained with Green viewer stain (CinnaGen, Iran), and the results were visualized under an ultraviolet (UV) light source using a GelDoc system (Genius3 Syngene, UK).
aThese primers consisted of the BsiWI (bolded in the forward 1 primer) and KpnI (bolded in the reverse 2 primer) restriction enzyme cleavage sites.
bTwo primers consisted of the overlapping part, bolded in the reverse 2 and forward primers.
PCR was performed to produce the first set of fragments. The PCR reaction mixture contained 5 μg of DNA sample (JHF1) (11), 1.5 mM of MgCl2 (CinnaGen, Iran), 0.8 mM of each dNTP (CinnaGen, Iran), 10 pmol of each primer (forward 0.8 and reverse 0.8 primers), and one unit of Taq DNA polymerase 5 u/µL (CinnaGen, Iran). Gene amplification was carried out using the following protocol: after an initial denaturation step of 95°C for 3 minutes, there were 28 cycles (95°C for 30 seconds, 59°C for 45 seconds, 72°C for 30 seconds), and a final annealing step of 72°C for 7 minutes. The second fragment’s PCR reaction mixture contained 1 μg of DNA sample (JHF1), 3 mM of MgCl2 (CinnaGen, Iran), 0.3 mM of each dNTP (CinnaGen, Iran), 10 pmol of each primer (forward 2 and reverse 2 primers, Table 1), and one unit of Taq DNA polymerase 5 u/µL (CinnaGen, Iran). After an initial denaturation step of 95°C for 4 minutes, there were an additional 30 cycles (93°C for 1 minute, 58°C for 30 seconds, 72°C for 50 sec), and a final annealing step of 72°C for 3 minutes. In the final PCR round, the overlap-PCR reaction was performed using 7 μL of the first and second PCR products (approximately 5 μg of template DNA of each product), 2.5 mM of MgCl2, 0.4 mM of each dNTP, and two units of Taq DNA polymerase 5 u/µL (CinnaGen, Iran).
The first round of gene amplification was carried out for 5 cycles (95°C for 1 minute, 65°C for 50 seconds, 72°C for 45 seconds), followed by the addition of 0.8 μL of 10 pmol primers (forward 1 and reverse 2 primers, Table 1). The second round of amplification was performed for 28 cycles (95°C for 40 seconds, 60°C for 50 seconds, 72°C for 45 seconds), and a final extension of 72°C for 3 minutes. The final PCR products (1645 bp) were purified using a commercial kit (Bioneer, South Korea) according to the manufacturer’s recommendations. The amplified igVR-1a chimera gene and JFH1 vector were digested using the KpnI and BsiWI restriction enzymes (New England Biolabs, UK) and the digested products were purified using a commercial kit (Bioneer, Korea) according to the manufacturer’s recommendations. The ligation mixture was prepared as follows: 0.8 µL of T4 DNA ligase (Fermentas, USA), 1 µL of 10× T4 DNA ligase buffer, 180 ng of purified linear JFH1 vector, 50 ng of purified igVR-1a chimera fragment, and 0.5 µl of 50% PEG 4000 solution. This mixture was prepared and incubated at 14°C overnight. The ligation product was transformed into E. coli JM109 strain competent cells, as described previously (12). The presence of the igVR-1a chimera clone was confirmed by colony PCR using the forward 1 and reverse 2 specific primers (Table 1) and a restriction enzyme analysis. The confirmed cloned fragments were used for purification with a commercial kit (Qiagen, USA) according to the manufacturer’s recommendations.
3.3. Sequencing
The cloned fragments were purified and sent for sequencing to Bioneer (Korea). The sequencing results were blasted.
3.4. In vitro RNA Transcription
The igVR-1a chimera gene, GND, and JFH1 vectors were digested with the XbaI restriction enzyme (Roche, USA). For in vitro RNA transcription, mung bean nuclease catalyst was used in the degradation of single-stranded DNA and RNA (Promega, USA). In vitro RNA transcription was performed with a MEGAscript T7 kit (Ambion, Germany) according to the manufacturer’s recommendations. After DNase I treatment, RNA was used for transfection (12).
3.5. Cell Culture
The necessary amount of the Huh7.5 cell line was received from the Pasteur institute of Iran. This cell line was grown in Dulbecco’s modified minimal essential medium (DMEM) (Gibco, USA) supplemented with 100 U/mL of penicillin, 100 µg/mL of streptomycin antibiotics (Gibco, Germany), 10% (v/v) heat-inactivated fetal bovine serum (Gibco, Germany), and 1 M of HEPES buffer solution (Invitrogen, USA). The Huh7.5 cells were incubated at 37°C in a 5% CO2 environment (12).
3.6. RNA Transfection and Infection of Huh7.5 Cells
The Huh7.5 cells were seeded at a density of 3 × 104 cells/well in 24-well plates, then cultured at 37°C overnight in DMEM medium. The confluency of cells reached 70%. RNA transfection was performed using DMRIE-C (Invitrogen, USA) in accordance with the manufacturer’s specifications. After transfection, the supernatants of the cultured cells were harvested and stored at -70°C. The Huh7.5 cells were seeded at a density of 3×104 cells/well in 24-well plates. Next, the cells were grown at 37°C for 24 hours in complete DMEM medium. As a result, the confluency of cells reached 60% - 70%. The supernatant of the previous step was transferred to the Huh7.5 cells. Finally, after 24 hours, the supernatants of the cultured cells were harvested and stored at -70°C.
3.7. Real-Time PCR of igVR-1a Chimera Virus
Total viral RNA was extracted from 140 µL of the stored supernatant of the cultured cells using a QIAamp viral RNA kit (Qiagen, USA) according to the manufacturer’s recommendations. Real-time PCR was performed using a commercial kit (Genome Diagnostics Pvt., New Delhi, India) (12).
3.8. Enzyme-Linked Immunosorbent Assay (ELISA)
HCV core antigen detection ELISA assay kits (XC-1000, USA) were used for igVR-1a chimeric virus detection. ELISA was performed for the core protein to evaluate the production of active virus. After transfection of RNA virus in Huh7.5 cells, all virus proteins were assembled and the active virus was produced.
3.9. IgVR-1a Chimera Virus Infection of Huh7.5 Cells
Supernatants of the igVR-1a chimera virus were used to infect Huh7.5 cell cultures. Cell-free supernatants were infected for 30 passages and then harvested and stored at -70°C.
3.10. Cloning and Sequencing
After 10 passages of viral RNA, the igVR-1a chimera virus was extracted. RNA concentration was measured using a bioanalyzer device (NanoDrop 2000, Germany). In order to eliminate the plasmid vector, DNase I (Fermentas, USA), was used. In addition, cDNA production was analyzed with a PCR kit (Fermentas, USA). PCR and standard cloning procedures for the PCDNA3.1 vector were performed with appropriate restriction sites in forward and reverse primers, where the HindIII and EcoRI (Roche, Germany) restriction enzymes were applied. The primer sequences were as follows: forward 1: 5’-AACTAAGCTTTGATGATGAACTGGTCG-3’, reverse 1: 5’-AACTGAATTC AAGCAACGCCAAAAGCC-3’ (fragment length 1720 bp). Next, PCR was performed using a thermal cycler (Applied Biosystems, USA). The final PCR product purification (1735 bp) and the final plasmid igVR-1a chimera colonies were performed using a commercial kit (Bioneer, South Korea). All plasmids of the igVR-1a chimera were then confirmed by colony PCR with forward and reverse specific primers, as well as sequencing (Macrogen, Korea).
4. Results
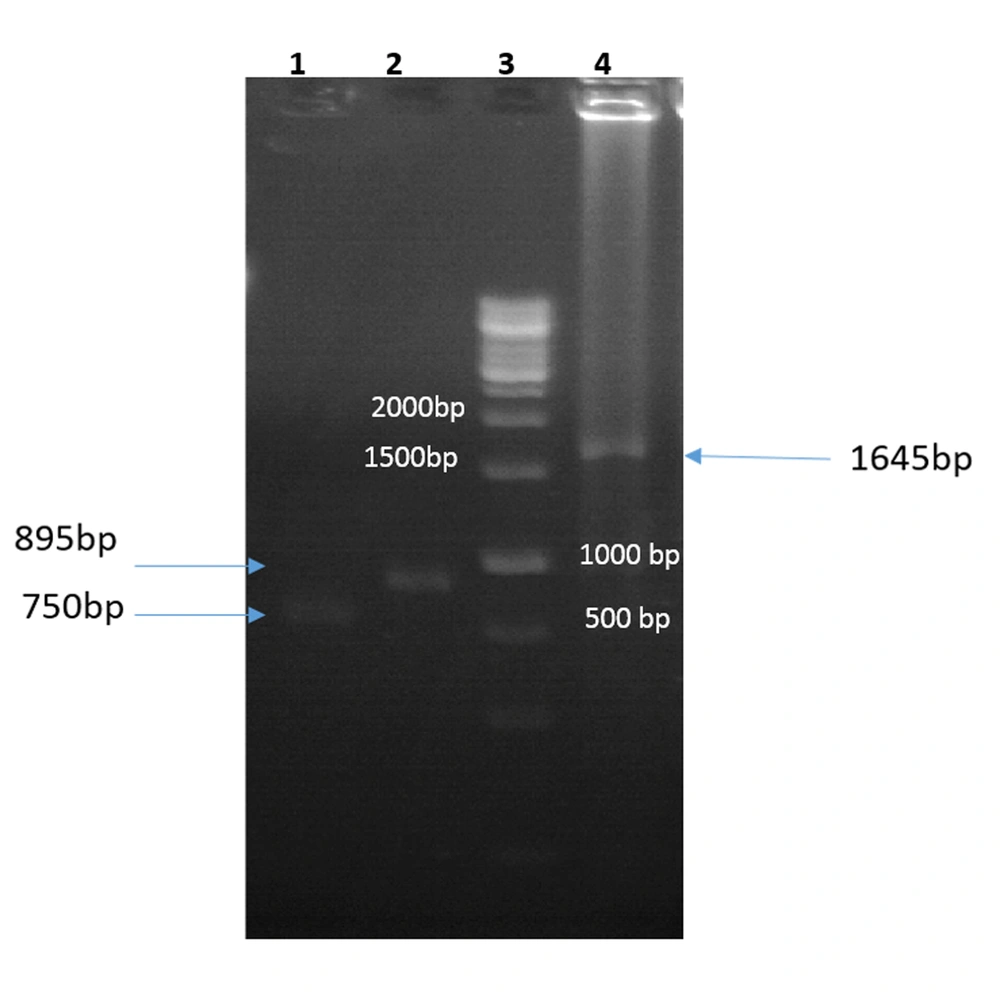
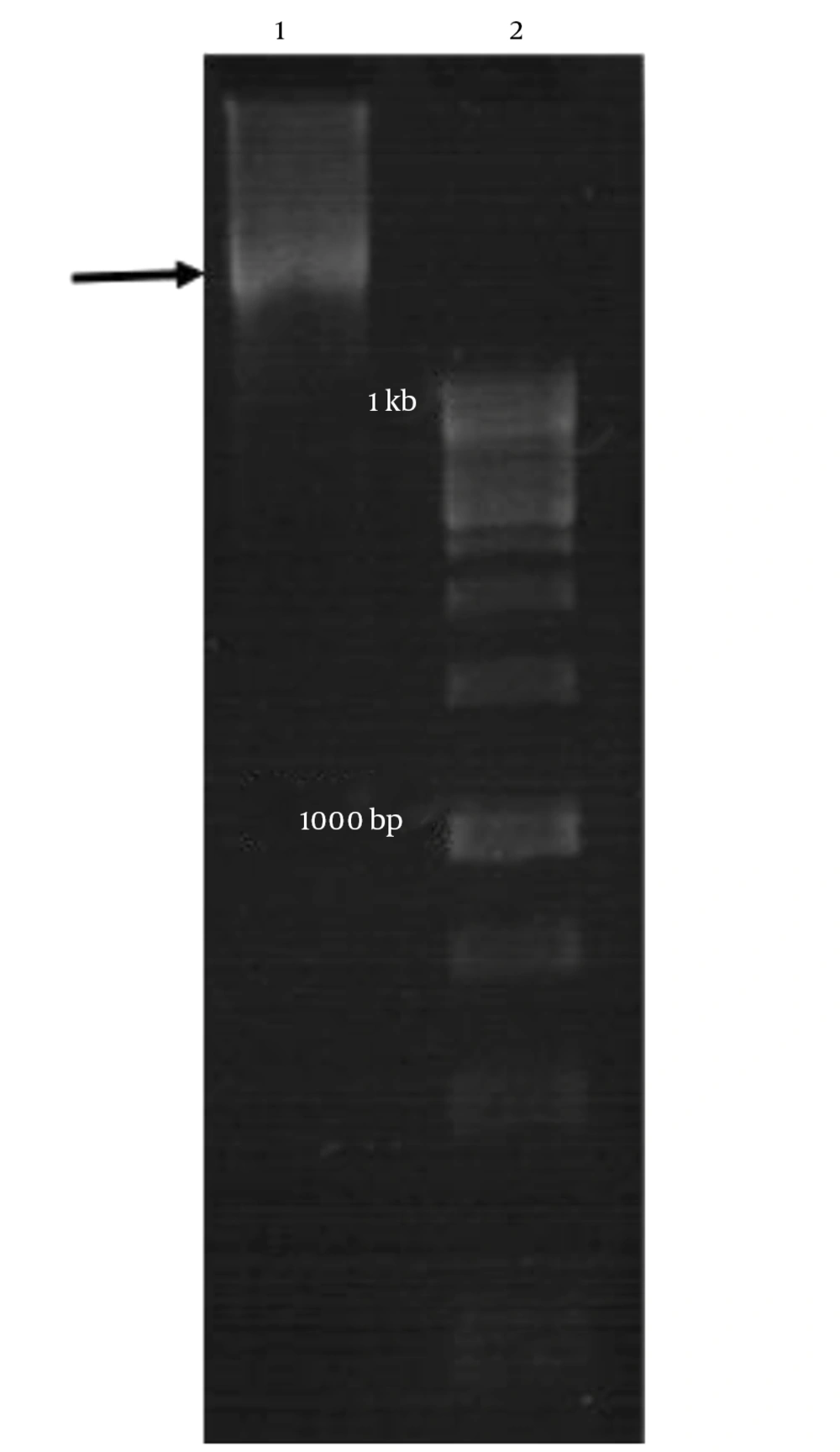
The igVR-1a chimera gene was amplified using the overlap-PCR method. The amplified fragments (1-2) and the final fragment had an expected size of 750 bp, 895 bp, and 1645 bp, respectively (Figure 1). After digestion with restriction enzymes, the DNA fragments containing the igVR-1a chimera gene were cloned into the JHF1 vector.
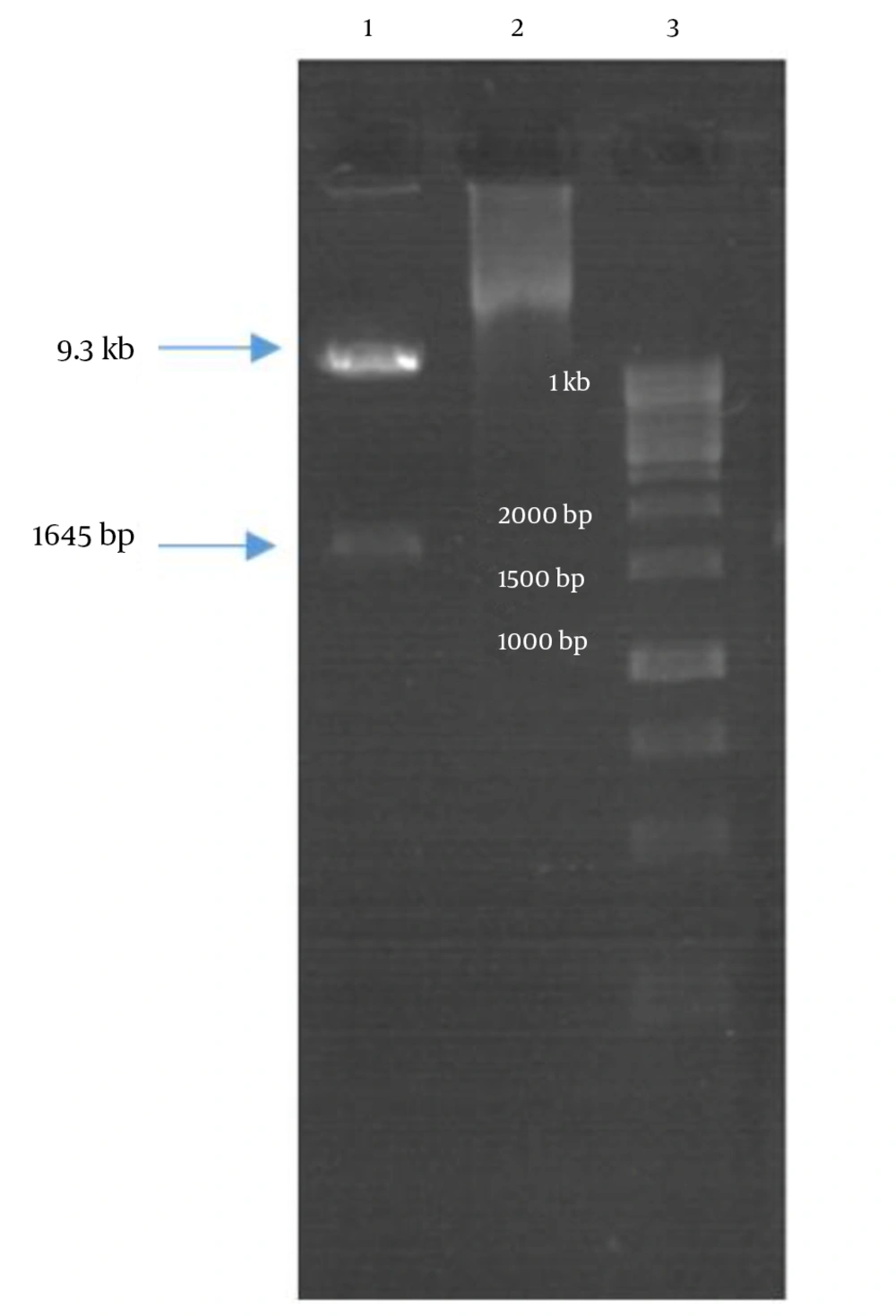
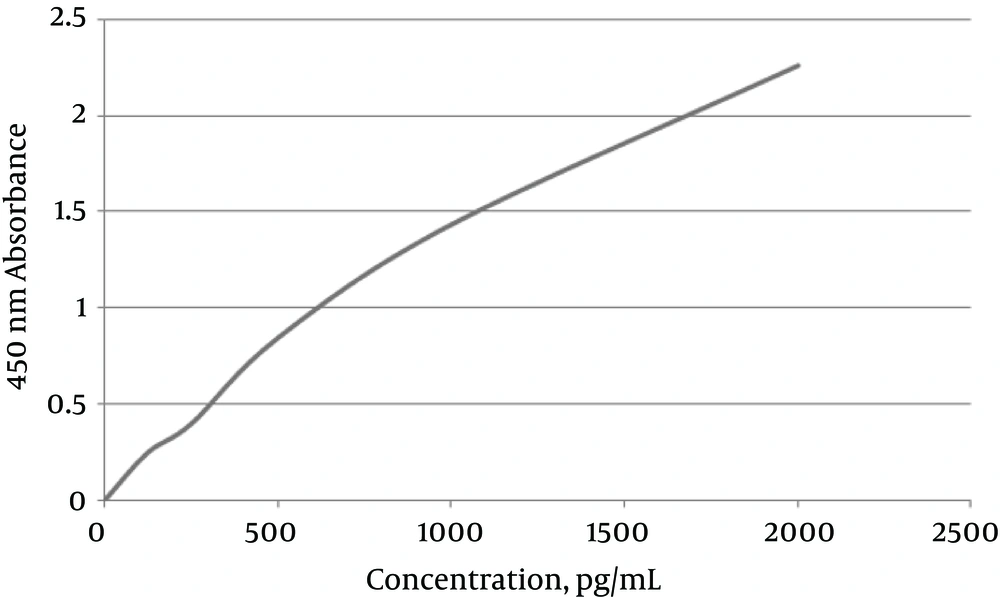
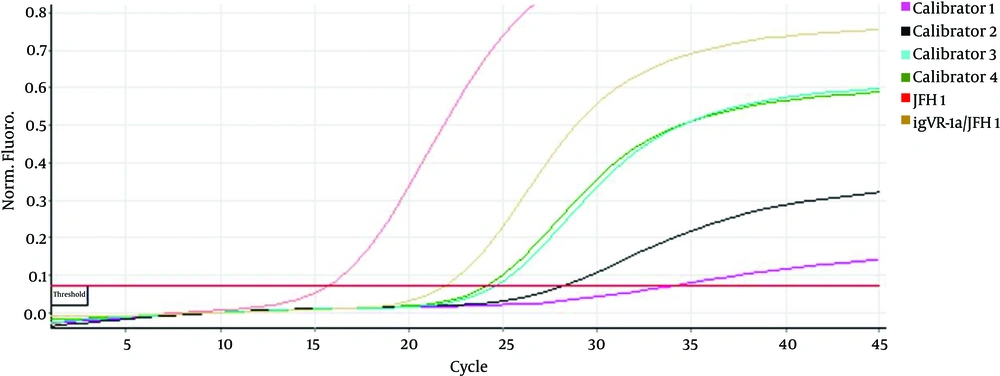
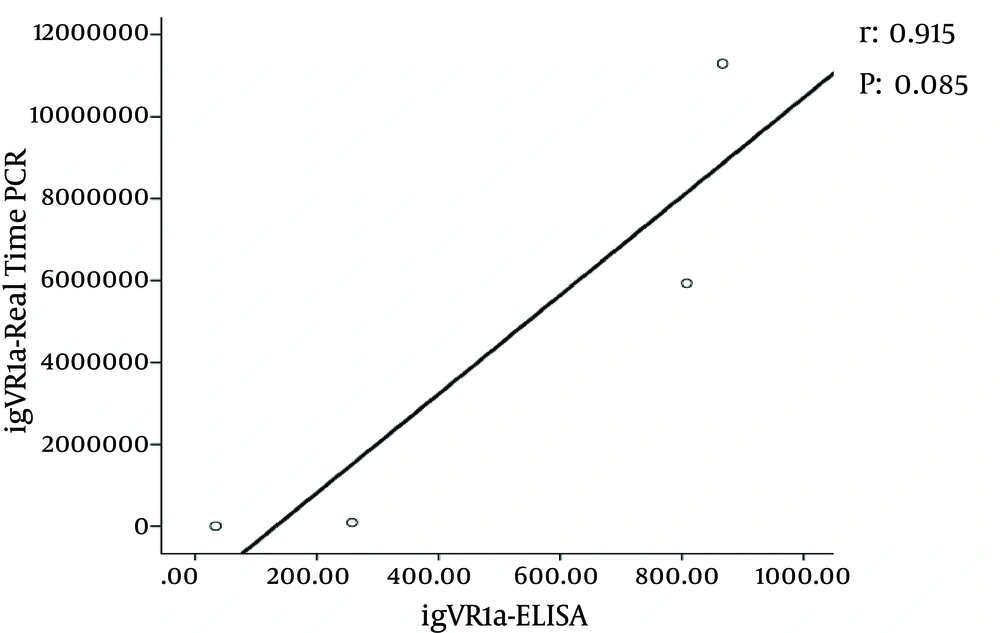
Double digestion of the igVR-1a chimera gene was done with the KpnI and BsiWI restriction enzymes (Figure 2). The igVR-1a chimera gene was digested with the XbaI restriction enzyme and in vitro RNA transcription (Figure 3). The standard curve of the ELISA assay kit for HCV core was used for igVR-1a chimeric virus detection (Figure 4). The igVR-1a chimera gene was cloned and purified. The results of sequencing showed that all fragments were 100% identical and had no gaps. After reaching almost identical RNA concentrations of igVR-1a and JFH1 in the same number of cultured cells, transfection was done. Next, virus protein production was performed using the ELISA method for evaluating the core protein of the virus. In order to examine the viability of the infectious virus, ELISA and real-time PCR were used after passages 1, 5, and 10. The ELISA report showed that JFH1 proliferated in passages 1, 5, and 10 (280.6, 367.6, and 621 pg/mL, respectively). Chimeric virus concentrations after passages 1, 5, and 10 were 34.5, 253.5, and 808.2 pg/mL, respectively. Real-time PCR results showed that JFH1 produced virus in passages 1, 5, 10, and 30 (5,017, 378,491, 12,615,815, and 13,390,851 copies/mL, respectively), while the chimeric virus titers after passages 1, 5, 10, and 30 were 6,040, 92,510, 5,933,449, and 11,292,514 copies/mL, respectively. Statistical analyses, such as correlation tests, were used to compare between the real-time PCR and ELISA results. There was no significant correlation between real-time PCR and ELISA results for igVR-1a (P > 0.05) (Figure 6). Correlation between real-time PCR and ELISA results was calculated with Pearson’s correlation test. Comparison of virus production during passages 1, 5, 10, and 30 with real-time PCR results in igVR-1a and JFH1 are shown in Figures 7 and 8. The GND plasmid as a negative control was not able to produce the virus. The construction of the igVR-1a chimera virus did not have any mutations, but an analysis of 30 passages of nucleic acids and amino acid sequences showed a change in their sequence.
5. Discussion
In the present study, we successfully developed a novel HCV chimera by substituting the igVR in the JFH1 subtype with that of genotype 1a. HCV is an enveloped, single-stranded RNA virus belonging to the Flaviviridae family (1). It is estimated that more than 3% of the world’s population is infected with HCV. Chronic HCV infections may predispose many patients to more serious liver disorders, including cirrhosis and hepatocellular carcinoma (3). The HCV genome is a circular RNA that encodes a large, 3,000-amino-acid polyprotein, which is cleaved onto different structural and non-structural proteins. Structural proteins include the core, the E1 and E2 glycoproteins, and a small p7 protein. The E1 (amino acids 193 - 383 of the polyprotein) and E2 (amino acids 384 - 746) glycoproteins of HCV are type I transmembrane proteins (13). The infectious viral particles of HCV (HCVpp) contain non-covalent E1E2 heterodimers. These viral particles are able to infect the primary human liver cells and various human hepatic cell lines, such as Huh7 (14). They are considered an alternative model for studying the virus-entry phase and entry inhibition. Glycoprotein E1 is pivotal for viral entry into the target cell; an HCVpp without it is not infectious (14). E2 contains 11 glycosylation sites and 18 conserved cysteine residues. Alignment of diverse E2 glycoprotein sequences across the six major genotypes revealed three variable regions between residues 570 and 580. HVR1, HVR2, and igVR are flexible structures in the surface region of the E2 protein that regulate the function of the E1E2 heterodimer and prevent the neutralizing antibodies from having access to the conserved and central constant region. Due to the presence of disulfide bonds, this region is protected against proteolytic activity. The variable regions of the E2 glycoprotein play an important role in the induction of the humoral immune response (14). The production of HCV chimeras has been an interesting branch of research since culturing of the JFH1 strain in the Huh7.5 cell line was first successful. Gottwein et al. succeeded in creating an intergenotypic 3a/2a chimera containing the core, E1, E2, p7, and NS2 of the 3a genotype, which was able to be cultured in Huh7.5 cells (15). Lu et al. also constructed a cultivable infectious intergenotypic 1b/JFH1 chimera containing the core, E1, E2, p7, and the first transmembrane domain of NS2 from genotype 1b (16). Yi et al. constructed chimeric genomes encoding the structural proteins of genotype 1a within the backbone of JFH1 (17). In the present study, we successfully created a novel HCV chimera by substituting the igVR in the JFH1 subtype with that of genotype 1a. Our results suggest that the generated chimera is cultivable, can produce RNA in cell cultures, and is able to assemble and produce infectious particles. These robust intergenotypic recombinant cell culture systems for HCV will provide opportunities to further study different HCV genotypes and evaluate related pharmacotherapeutics. Moreover, alongside creating cultivable chimeras, compensatory mutations essential for the in vitro production of an infectious virus was identified in the aforementioned studies. Pietschmann et al. created a series of intergenotypic and intragenotypic chimeric genomes allowing the production of infectious HCV genotype 1a, 1b, 2a, and 3a particles. Moreover, it was shown that a polyclonal IgG derived from a mouse immunized with a recombinant vaccine consisting of the E1 protein from genotype 1a was able to neutralize the infectivity of HCV genotype 2a, 1a, 1b, and 3a particles (18).
According to a previous study by Meshkat et al. (11), Iran has a high prevalence of HCV genotype 1. Unfortunately, this genotype cannot be cultured in vitro to analyze the behavior of the virus. On the other hand, the igVR of the virus’s surface glycoprotein E2 is highly effective at binding to the hepatocyte receptor. In this project, we exchanged this part of the virus with the same region of the JFH1 (genotype 2) glycoprotein E2 and created an active chimera virus in cell culture. We studied the structure and behavior of the virus and multiple mutations of glycoprotein E2, in order to investigate the survival, adaptation, and replication of the chimera virus in cell culture. After 30 passages, we observed changes in the amino acids of igVR-1a in the chimera virus, where Cys was exchanged with Val. Val was also exchanged with Thr, and Gly was replaced by Ala. The observed changes might result in alterations in the three-dimensional structure of the proteins, such as the changes in the enveloped E2 glycoprotein on the surface of HCV. These variations affected the binding of HCV with cell receptors and therefore resulted in changes in the pathogenesis of HCV.
The aim of this project was to produce a chimera virus as a combination of genotypes 1a and 2a. The 1a genotype has a high prevalence in Iran and the 2a (JFH1) genotype has the ability to be cultured in vitro. Our objective was to evaluate the structure of the nucleic acids and amino acids of the chimera virus. We replaced the igVR between the two genotypes to produce HCVpp. Next, we analyzed this chimera virus with ELISA and real-time PCR. Chimeric HCV-1a produced additive concentrations in cell-culture systems and the novel HCV chimera in all passages in the cell cultures, such as JFH1. These novel intergenotypic recombinant viruses, using the backbone of the JFH1 virus and the igVR of genotype 1a, were tested in an animal model and the immune responses were analyzed. The results can be used for reverse genetic studies and research into neutralizing antibodies, virus assembly, and vaccine development.
5.1. Conclusion
In the present study, we successfully created a novel HCV chimera using the backbone of the JFH1 virus and the igVR of genotype 1a. Our chimeric virus proliferated in Huh7.5 cell cultures and produced infectious viral particles. We substituted the igVR domain of the JFH1 virus with that of genotype 1a, which resulted in the production of viral particles. A novel HCV chimera, using the backbone of the JFH1 virus and the igVR of genotype 1a, can be used for simulating an in vivo animal model and for analyzing the immune response in future reverse-genetic studies.