1. Background
The hepatitis B virus (HBV) is an enveloped virus with an outer diameter of approximately 42 nm contained within a nucleocapsid. Capsids enclose a single copy of the 3.2-kb, partially double-stranded DNA genome, which is covalently linked to the viral polymerase at the 5’ end of the full-length minus strand. Coding regions in the HBV genome are organized into four overlapping reading frames (ORFs) designated C (core), P (polymerase), S (surface), and X (a regulatory protein), which are subsequently translated into the corresponding viral proteins (1).
The HBV envelope proteins can be translated from a single ORF: L (large), M (middle), and S (small) or the hepatitis B surface antigen (HBsAg) (1). The HBsAg is composed of four transmembrane helices that are involved in the integration of protein into the endoplasmic reticulum (ER) membrane. Other regions of the HBsAg are highly coiled, and the coil is responsible for protein’s antigenicity (2). There is a major hydrophilic region (MHR) that encompasses amino acid residues 99 and 160, which contains the major epitopes for the induction of a humoral immune response (3, 4). The “a” determinant domain (amino acid positions 121 - 147), which is a highly conserved region of the HBsAg, is located on the exterior surface of the MHR and is involved in the binding of antibodies (anti-HBs) against HBsAg (4, 5). Several HBV mutations within the ”a” determinant of the HBsAg have been reported as immune escape mutations, which can potentially be involved in vaccine-induced immunity and diagnostic-escape variants (4). The commonest type of these mutations, G145R, is created by the substitution of arginine for glycine has been shown to exhibit various degrees of altered binding of HBsAg to antibodies in different commercial assays (6, 7). G145R mutant has been reported in many cases of occult hepatitis B infection (OBI) because it decreases the HBsAg levels (8, 9), often going undetected by routine assays (10) and in patients who suffer from lamivudine-resistant mutants (11). HBV has been classified into eight genotypes (A - H) based on sequence divergence in the genome (12). G145R mutants have mainly been found in genotypes B, C, and D (13). Naturally occurring G145R mutants are often detectable with monoclonal antibody-based assays, albeit at a reduced sensitivity (14).
Previous studies have indicated that the “a” determinant region interacts with the antibodies from patient serum (15) or the mouse monoclonal antibody produced against HBsAg (16). One of the obstacles to detecting a variant critically depends on the choice of the antibody. In contrast, the fundamental difficulty in the in vitro characterization of all this variation is the difficulty in the quantitation of the expressed HBsAg in a way that does not depend on its antigenicity. One approach involves the use of an antibody that binds to a common region away from the variant domains being tested, but how can one be sure that the structural conformation is not affected? Moreover, due to a lack of crystallization of wild-type HBsAg molecules and membrane-spanning (17, 18), no template structure exists in the protein data bank (PDB) library for the HBsAg (19).
2. Objectives
The objectives of this de novo study were to assess the impact of the G145R mutation on the HBsAg structure at both the two-dimensional (2D) and three-dimensional (3D) levels. We also performed molecular docking studies of the HBsAg-antibody to investigate the antigen-antibody interactions in the G145R mutant compared with the wild-type HBsAg.
3. Materials and Methods
3.1. 2D Analysis and Transmembrane Topology
Predictions regarding the secondary structures of both wild-type (accession numbers: GQ183486) and G145R mutant HBsAg were carried out using three highly accurate secondary structure prediction tools: Jpred 4, PHD, and PSIPRED (20-22). Three different servers were used to elevate the accuracy of the predictions. In parallel, the MEMSAT3 and TMpred web tools (http://embnet.vital-it.ch/software/TMPRED_form.html) were also applied to predict the orientation and topology of the transmembrane regions of the G145R mutant compared with the wild-type HBsAg (23).
3.2. 3D Analysis and Structural Alignment
Using the amino acid sequence of the HBsAg, a PSI-blast was performed to determine the most suitable homologous structures. The results indicated that there was no appropriate template structure for homology modeling; therefore, we used a threading method for the HBsAg model’s construction. A Phyre2 server was applied to predict the 3D structure of the HBsAg; this server utilizes the alignment of hidden Markov models using an HMM-HMM comparison, which can significantly improve the accuracy of the alignment and detection rates. It also includes a new antibody initio folding simulation called Poing, for multi-template modeling and antibody initio protein structure prediction (24). Finally, a model was generated by Phyre2 and then subsequently refined using the ModRefiner (25). After several energy minimization steps using GROMACS 43a1 force field, the visualization of the refined model as well as the generation of the G145R mutant were performed by PyMOL software (26). To quantitatively evaluate the modeled 3D structure, several web tools, including VADAR, PROSESS (http://www.prosess.ca/), and SAVES (nihserver.mbi.ucla.edu/SAVES/), were used simultaneously (27). At the same time, the wild-type and G145R mutant HBsAg proteins were structurally aligned, and their root mean square deviation (RMSD) and template modeling (TM) scores were computed using the SuperPose and TM-align servers, respectively (28, 29).
3.3. Molecular Docking
It is now well established that the monoclonal antibody 12 (MAb12) can selectively bind to the surface antigen of the hepatitis B virus (30). The 3D structure of MAb12 was obtained from the PDB database (http://www.rcsb.org/pdb; PDB ID: 4Q0X). Unwanted ligands, ions, and molecules were removed from the PDB file using PyMOL software, and the final refined MAb12 structure was used in molecular docking studies. Molecular docking of the HBsAg and MAb12 structures was performed using the ClusPro web server (31). The binding affinity of the HBsAg-MAb12 complex was calculated according to the coefficient weights as follows:
E = 0.20Erep + - 0.40Eatt + 600Eelec + 0.25EDARS
Where Erep and Eatt represent the repulsive and attractive components in the implementation steps of the van der Waals energy, respectively. Eelec is the Coulombic electrostatic energy, and EDARS denotes a new class of knowledge-based interaction potentials. See Brenke et al. paper for more details (32).
3.4. Molecular Dynamics (MD) Simulations
MD simulation has been widely used in structural biology investigations and can serve as a powerful tool to determine the atomic-level behavior of biological events, such as protein conformational changes under different physiological conditions (33). MD simulations for both the G145R mutant and the wild-type HBsAg were conducted using GROMACS v5.0.2 package (34). The predicted 3D structures were solvated in a solvation box with 10Å distance between the edges of the box and the protein fragments. Na+ and Cl- ions were added to neutralize the system. Then, the equilibrated systems were subjected to a 10-ns MD simulation in the isothermal-isobaric (NPT) ensemble using the leap-frog algorithm with an integration time step of 0.002 ps.
3.5. Evaluation of Protein Antigenicity
It has been sufficiently established that amino acid substitutions in both within and outside of the “a” determinant can cause a reduced binding affinity to monoclonal antibodies (35). To assess the antigenic potential, both wild-type and G145R mutant HBsAgs were subjected to the antibody epitope prediction server IEDB-AR (36), which provides multiple tools to predict MHC class I and MHC class II restricted T-cell epitopes as well as linear and discontinuous B-cell epitopes.
4. Results
4.1. G145R Mutation Alters the Secondary Structure and Membrane Orientation of the HBsAg
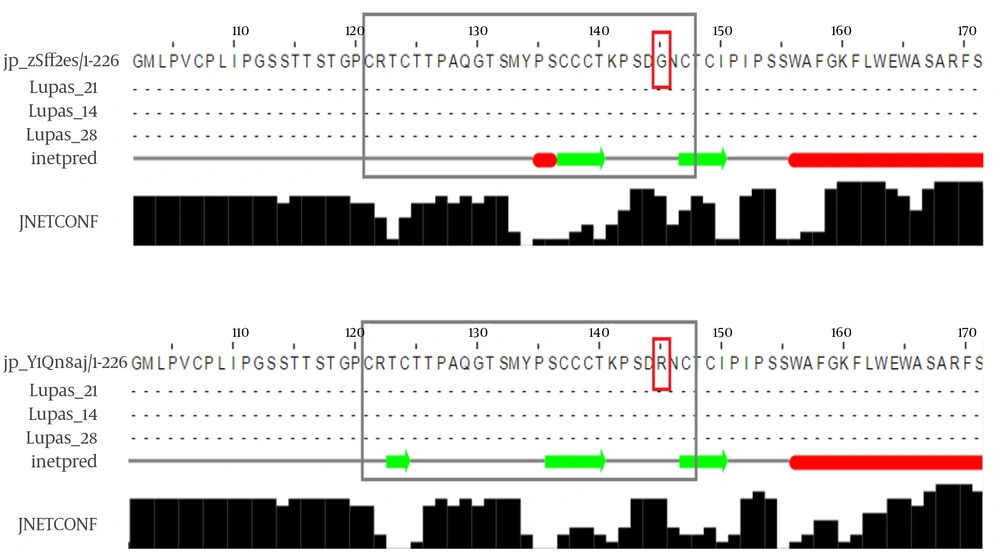
G145 is located in the second loop of the MHR within the “a” determinant, a flexible coil stabilized by two disulfide bonds that play a key role in the antigenicity of HBsAg. A secondary structure analysis of the G145R mutant revealed that this substitution inserted a new β-strand at the 121 - 124 position (Figure 1). However, the differences between the secondary structure of the G145R mutant and the wild-type HBsAg at all other regions of the protein were extensively subtle.
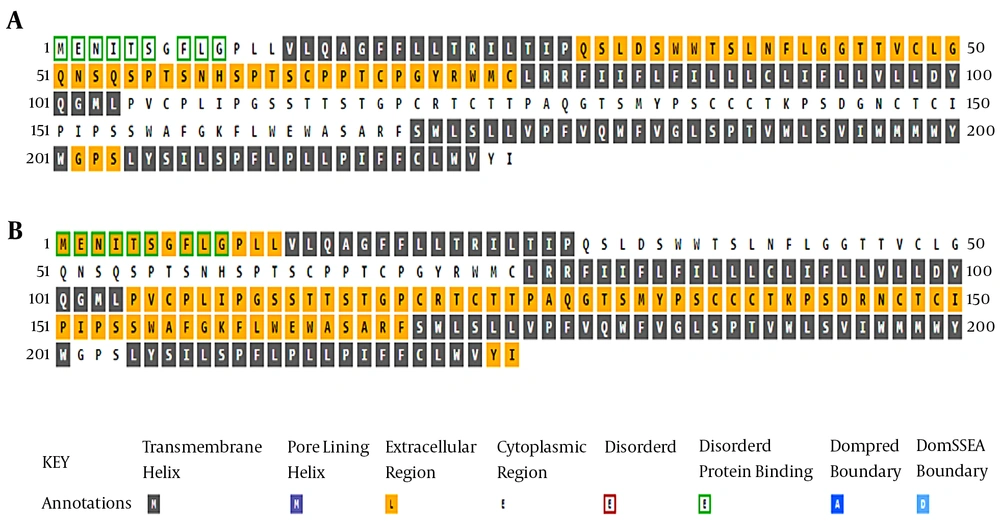
An analysis of the transmembrane regions revealed that the number of transmembrane helices in the G145R mutant was not altered compared to the wild-type. However, the orientation of transmembrane segments between the G145R mutant and wild-type was clearly different, suggesting the possible impact of the G145R mutation on the altered localization of the mutant HBsAg within the ER membrane. The “a” determinant region in the wild-type and G145R mutants is positioned at the ER lumen and in the cytoplasmic area, respectively (Figure 2). Accordingly, any alteration in the membrane topology of the HBsAg can affect the distribution of this protein.
4.2. Prediction and Evaluation of the 3D Structure of the HBsAg
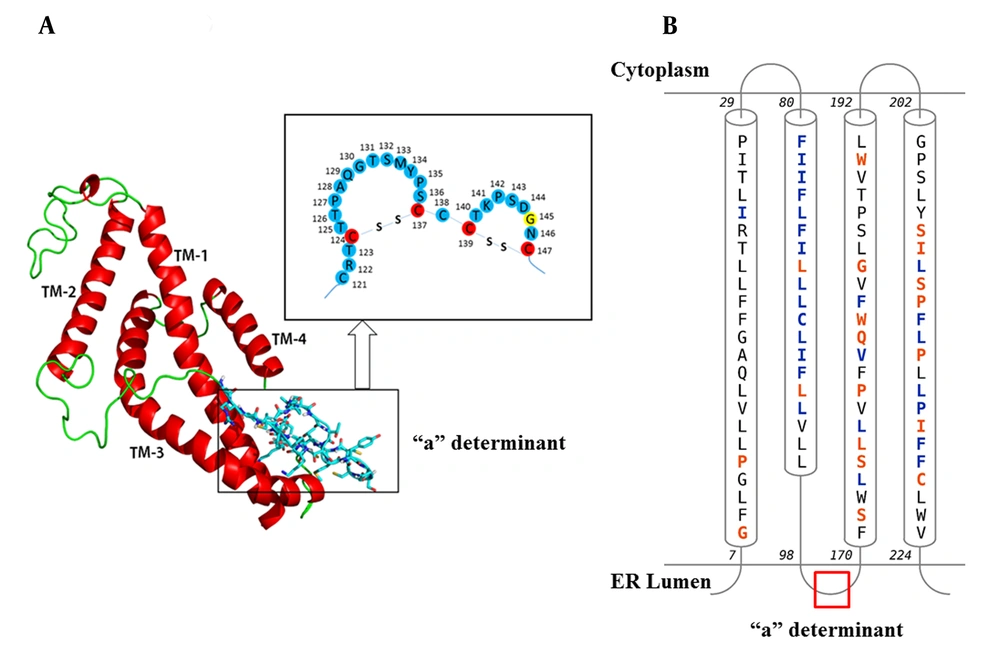
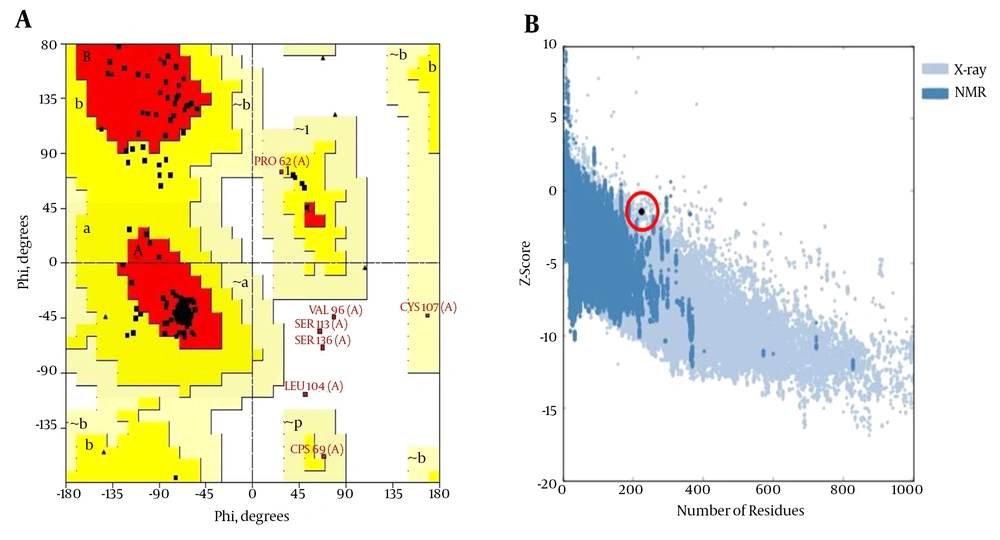
The predicted 3D structure of the HBsAg after the refining and energy minimization steps is indicated in Figure 3. A Ramachandran plot of the final refined HBsAg model obtained from PROCHECK revealed that 87.7%, 9.1%, 1.1%, and 2.1% of the residues were located in the most favored, additionally allowed, generously allowed, and disallowed regions, respectively (Figure 4A). The overall model quality was evaluated using the ProSA Z-score, which calculates the overall quality of the model to determine whether the quality of the input model is positioned within the range of scores typically found for experimentally solved protein structures (37). The ProSA Z-score for the final HBsAg model was -1.39, which falls within the normal range of experimentally validated protein structures (Figure 4B).
A) The Predicted 3D Structure of the HBsAg Along with a Schematic Representation of the “a” Determinant Region. Transmembrane helices (TM1-4) are shown in red and the mutation site (amino acid 145) is yellow; B, Topology of HBsAg helices within the ER Membrane. Contact Colorcodes: Orange and Blue Letters Refer to Helix Contact and Membrane Contact, Respectively.
The residues in the most favored (red), additionally allowed (yellow), generously allowed (pale yellow), and disallowed regions (white) of the Ramachandran plot are indicated. The structures determined by different methods (X-ray, NMR) are distinguished by different colors in the ProSA Z-score plot.
Further quality analysis was carried out using the MolProbity score. This score for the modeled 3D structure of the HBsAg was calculated as 1.94, which indicates a good resolution of this model. The MolProbity is derived from a combination of the clash score, rotamer, and Ramachandran evaluations. In addition, the MolProbity clash score was 8.4 (the number of serious steric overlaps (0.4 A°) per 1000 atoms). Structures with a high quality usually score below 10. 3D verification was allocated to determine the compatibility of the HBsAg 3D model with its own amino acid sequences. The results indicated that 69.47% of the residues were scored 0.2. The outcome is considered acceptable when more than 65% of the residues are 0.2. The predicted structure was further validated using ERRAT, which analyzes the statistics of non-bonded interactions between different atom types. A higher ERRAT value suggests a higher quality 3D model. The ERRAT score for the HBsAg model was 76.09%, which was satisfactory; this finding was also in agreement with previous overall quality scores.
4.3. The Effect of G145R Substitution on the 3D Structure of the HBsAg
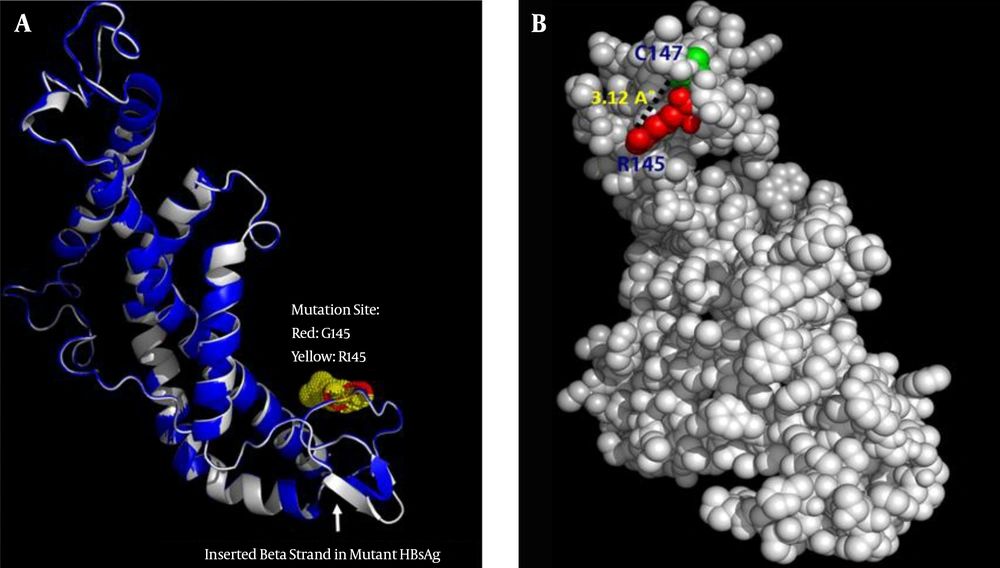
Although the superposition of two structures demonstrates an inconsiderable alteration in the overall 3D structure, this mutation can induce local changes in the structure of the protein, particularly in the “a” determinant region. Figure 5A represents the structural alignment of the G145R mutant and wild-type HBsAg. Some of these changes, such as the number of residues with an H-bond, the total accessible surface area, and the total volume, were computed using the VADAR web server. Compared to the G145 residue, the R145 can interfere with the integrity of the HBsAg antigenic loop by forming an H-bond with C147, a key residue involved in the formation of the second antigenic loop of the HBsAg (Figure 5B). The very low RMSD value (0.38) and the high TM-score (0.99) that resulted from the structural alignment demonstrate that the G145R mutant and wild-type HBsAg have almost the same folding character.
A, Superposition of the wild-type HBsAg (blue) and the G145R mutant (gray). The arrow indicates the position of the inserted β-strand in the mutant HBsAg’s “a” determinant region. B, Representative positioning of the H-bond between the R145 and C147 mutation in the 3D structure of the G145R mutant HBsAg.
4.4. G145R Mutation Decreases the Binding Affinity of the HBsAg to the MAb12
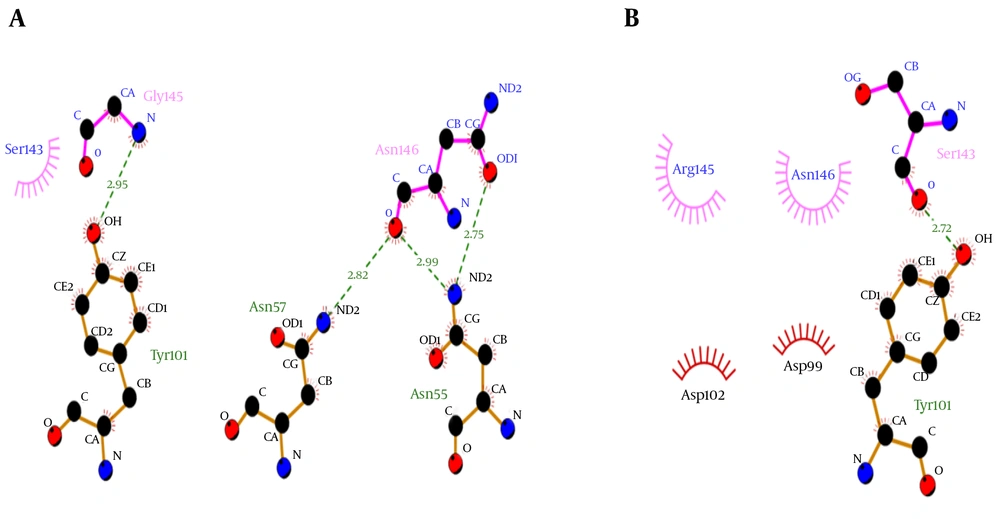
Molecular docking showed that the G145R mutation caused a noticeable decreased binding affinity between the HBsAg and MAb12. The wild-type and G145R mutant HBsAgs were bound to the MAb12 with weighted scores of -626.7 and -309.1, respectively, implicating the lower binding affinity of G145R mutant to MAb12 compared to wild type. The HBsAg-MAb12 complexes were visualized using the DIMPLOT module of the LigPlot+ package to determine their polar bonds and non-polar connections (38). The results demonstrated that the G145R mutation likely reduced the number of H-bonds through residue-residue approximation constraints with adjacent amino acids. Furthermore, a DIMPLOT analysis indicated that G145 and N146 were actively involved in the HBsAg-MAb12 interaction. In the mutant HBsAg, R145 did not form any H-bond with MAb12. The larger size of the R145 side chain seems to be involved in the inhibition of H-bond formation between the N146 and MAb12 residues (Figure 6). More details regarding the molecular docking results are shown in Table 1.
| MAb12 | ||||
|---|---|---|---|---|
| HBsAg type | Center Energy | Lowest Energy | Cluster No.a | “a” Determinant H-Bond No. |
| Wild-type HBsAg | -601.8 | -626.7 | 177 | 14 |
| G145R mutant HBsAg | -290.9 | -309.1 | 66 | 10 |
aThe cluster with the largest number of low-energy structures typically contains the native fold.
4.5. G145R Mutation Reduces the Antigenic Potential of the HBsAg
An evaluation of the protein antigenicity revealed that the antigenic potential of G145R mutant HBsAg was reduced compared to the wild-type (Figure 7). The antigenicity scores for the G145 and R145 residues were calculated as 1.19 and 0.9, respectively, implicating a key role for G145 in the antigenic potential of the HBsAg.
4.6. MD Simulations
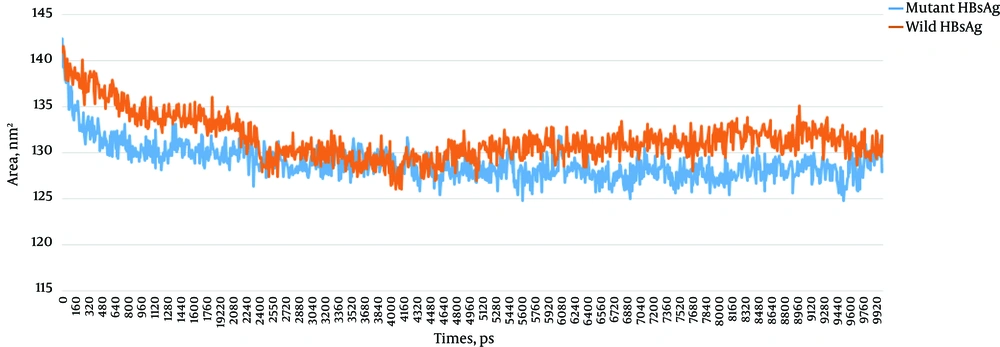
RMSD is a reliable value that is widely used for evaluating the structural differences between proteins and is also considered to be an indicator of structural stability. The RMSD plots of the wild-type and G145R mutant HBsAg revealed that the G145R mutation further stabilized the HBsAg’s structure (Figure 8A). We also calculated the radius of gyration (Rg) values for both HBsAg types (Figure 8B). The Rg is most often used in the determination of protein compactness (39). A comparison of the Rg plots demonstrated a higher compactness of the mutant HBsAg compared to the wild-type HBsAg.
To determine the structural dynamics of the wild-type and G145R mutant, the root mean square fluctuation (RMSF) of the Cα of the HBsAg protein was compared between the trajectories generated at 300 K to gain an insight into the flexible nature of the G145R mutant compared with the wild-type. Interestingly, the flexibility of the mutant HBsAg, especially in the antigenic regions, was considerably reduced, indicating that the G145R mutation resulted in increased protein rigidity (Figure 9), which makes the HBsAg unsuitable for interacting with antibodies.
A calculation of the accessibility surface area (ASA) showed that the surface accessibility of the G145R mutant HBsAg was reduced upon G145R substitution. ASA plots of the wild-type and mutant HBsAg are shown in Figure 10. A decrease in the mutant HBsAg ASA suggests that the G145R mutation converts the HBsAg structure to a compact configuration with buried functional segments, such as epitopes. The ASA of the side chains in the wild-type and the G145R mutant were 13,219.2 and 13,017.8 Å 2, respectively.
5. Discussion
A variety of HBsAg mutations has been reported within the “a” determinant region (4). Of these, G145R is the commonest and the most widely studied HBV immune escape mutation, emerging naturally or in response to vaccination or hepatitis B immunoglobulin (HBIG) therapy. Previous studies have indicated that the ability to neutralize anti-HBs against this variant could be diminished (40-42). In the current study, we investigated the 3D structure and molecular dynamics of the HBsAg at position 145 for both the wild-type and the mutant. Amino acid mutations within the critical positions of structural proteins may influence its stability and conformation and also affect its properties and interactions with other proteins. Several studies have indicated that the 3D structures of proteins play a pivotal role in their biological activity and interactions with other molecules (43).
In this study, we found that the secondary structure of the G145R HBsAg was significantly affected, with a gain of a β-strand as well as the loss of α-helix contents. It is now widely accepted that β-strands play a key role in the aggregation of proteins. The aggregates have more β-strand structures and less α-helix content (44). Taken together, the G145R mutation can increase the rigidity and aggregation potential of HBsAg’s “a” determinant, which alters its immunogenic activity and secretion. Recent studies have shown that the antigenic determinants of the HBsAg are highly conformation-dependent; therefore, any alteration in the “a” determinate region can reduce or inactivate the antigenic activity of this antigen (45).
Moreover, our results demonstrated that the G145R mutation was highly capable of increasing the compactness of the HBsAg. The compactness of a protein is generally described as the ratio of the ASA of a protein to the surface area of the ideal sphere of the same volume (39). These results confirmed the relationship between protein compactness and surface accessibility. Indeed, we found that the G145R mutation reduced the ASA of the HBsAg by increasing the protein’s compactness and rigidity. An increase in protein compactness is one explanation for the increased protein rigidity that leads to enhanced protein stability (46). It is plausible that a reduced protein ASA also may elevate the protein’s stability (47). Furthermore, an analysis of Rg demonstrated that the atoms of the G145R mutant were highly packed within the protein structure, resulting in increased protein stability. In contrast with the wild-type HBsAg, the RMSD of the coordinates of all Cα atoms in the G145R mutant demonstrated that the HBsAg mutant remained very stable and folded over the course of 10 ns at 300 K. The RMSD and Rg values are highly reliable measures for determining a protein’s compactness and stability under physiological conditions (48, 49). There is also a correlation between the Cα RMSF measures and protein flexibility; therefore, proteins with higher RMSF values are generally considered to have more flexible regions (50). The present study showed that the average Cα RMSF of the G145R mutant was clearly lower than that of the wild-type, especially in the antigenic regions. This finding may be an important factor involved in the antibody escape of HBsAgs that contain the G145R mutation (51).
An antigenic potential analysis of the wild-type HBsAg and the G145R mutant revealed a significant decrease of antigenicity in the mutant type. This finding was supported by other results from molecular dynamic and docking studies. The lower binding affinity between G145R and MAb12 compared with the wild-type protein indicated the intrinsic potential of this mutant to escape HBsAg neutralization using specific antibodies, as reported in previous studies (52-54). According to the detailed figures of amino acid interactions revealed by DIMPLOT, the substitution of Gly with Arg leads to a reduction in the total number of H-bonds between the HBsAg and its monoclonal antibody. Protein-protein docking results showed that unlike G145, R145 was not able to form an H-bond with MAb12. Additionally, it prevents the appropriate interaction of N146 with its target residues in MAb12. Therefore, R145 may change the arrangement of the H-bond network in the antigenic loops of the G145R mutant HBsAg. Moreover, when G145 is replaced by R145, a new H-bond forms between this residue and C147, which may also alter the spatial conformation of the second loop of the “a” determinant region.
Molecular dynamic analysis revealed a significant increase in the compactness of the mutant protein as well as a decrease in the flexibility of the antigenic loops and the total accessible surface area of the mutant type. All these results indicate that the antigenic loops in the G145R mutant are not as exposed as those in the wild-type HBsAg, and it can reduce its availability to specific HBsAg antibodies, which may explain the lower immunogenic potential of the G145R mutant protein. Reduced immunogenicity in “a” determinant mutants, especially the G145R mutant, has been observed in previous studies (13, 55). These mutations can specifically change the binding affinity to antibody as well as the sensitivity of diagnostic tests. Glycine is an amino acid with a non-polar side chain, while Arginine has a polar, charged (at physiological pH), and highly bulky side chain. Thus, differences in the physicochemical properties of these residues may affect the 3D conformation and therefore the steric hindrance between other residues (56).
The G145R mutation is an immune escape mutation that causes problems with diagnostic assays and a poor response to vaccines. Mutants can replicate, even in vaccinated individuals, so current vaccines are often ineffective in the production of suitable antibodies (54, 57, 58). Therefore, these mutants and their subsequent alterations in the HBsAg structure and characteristics deserve significant attention to facilitate the design of more specific antibodies for vaccines and serology designations.
In conclusion, this study provides some in silico evidence about the impacts of the G145R mutation on the structure and immunogenic activity of the HBsAg. Our analyses revealed that the G145R mutation induces a local change in the “a” determinant conformation. The results suggest that the HBsAg can escape from the host’s immune system, vaccine, and HBIG as well as from diagnostic assays due to a local change in the “a” determinant region. However, the results need to be validated experimentally.