1. Background
Chronic hepatitis B is an infectious disease caused by hepatitis B virus (HBV), which results in acute or chronic liver diseases. HBV infection is a public health problem. It is estimated that more than 350 million people worldwide are infected by this virus and 75% of them are Asians. Several evidence indicate that the clinical appearance of infection varies among individuals and includes the following modes: spontaneously recovered infection, chronic and acute HBV, cirrhosis or hepatocellular carcinoma, and liver cancer-related death (1).
Several factors such as characteristics of the virus, environmental factors, and host properties such as age, ethnicity, and genetic background can affect the susceptibility to the chronic HBV infection (2).
A study on twins in China showed that identical twins might have more incidence of HBeAg compare to the non-identical twins. It means that, the incidence of infection is influenced by host genetic factors (3). Moreover, reports indicate that genetic factors might increase the chronic HBV carrier in some populations (4).
The real mechanism of HBV infection is yet unknown. The studies about the host immune responses indicated a significant association between the T-cell functions and infectious diseases (5). More recently, results suggest that chronic HBV infection is an immune related disease, and cellular immune activation was observed through some cytokines such as interleukin 10 (IL-10) (6) and interleukin 28 (IL-28) (7, 8), as well as macrophage migration factor (9). Cytokines play an effective role in host immune system. These compounds can reduce the viral replication and control the host immune response. In addition, the host genetic background also regulates the natural history of chronic HBV infection (10). Based on the relationship between cytokines and immune responses, the serum level of cytokines has an effect on disease outcome (11). In this regard, single nucleotide polymorphisms (SNPs), specially in promoter and regulatory regions of the cytokine genes, change the level of cytokine expression and host response, as well (12).
Tumour necrosis factor-alpha (TNF-α) is a potential pro-inflammatory cytokine, which increases in HBV-infected individuals and induces the clearance of HBV via reducing the replication of HBV in the hepatocytes (13, 14). In-vivo studies indicated that TNF-α can reduce the existence of HBV antigen via the regulation of the human leucocyte antigen (HLA)-II molecules (15, 16). Also, TNF-α levels vary in subjects with some SNPs in promoter regions (17-19).
Human TNF-α gene is located on chromosome 6p21.3 and has 4 exons. Nowadays, more than 600 SNPs in the TNF-α gene have been identified, which some of them (-238 G/A, 244 A/G, 308 G/A, 376 A/G, 575 A/G, 857 C/T, 863 C/A, 1031 T/C, 1125 G/C, and 1196 C/T) are located in the promoter (20) and can change the production of protein (21). Velayati et al. (22) investigated the association of TNF- α gene polymorphisms among Iranian pulmonary tuberculosis patients as an infectious disease. They found that polymorphisms in TNF-α-238 gene were associated with the risk of developing pulmonary tuberculosis. Studies in Caucasians revealed a positive correlation between -238A allele and chronic HBV infection (23). In the Korean patients, the resolution of HBV infection was associated with the -308A allele and -863 CC genotype. Also 163G/238G/308G/857C/863C/1031T and 163G/238G/308G/857C/863A/1031C haplotypes were related to the clearance and persistence of infection, respectively (24). Fidder et al. (25) reported that -1031 C, -308 A, and -238 A alleles are related to the increased levels of TNF-α, while the -863 A allele is related to the reduced level of TNF-α. In above mentioned studies, conflicting results have been reported, hence further studies are necessary.
2. Objectives
In this case-control study, we aimed to explore whether the TNF-α promoter SNPs (-238 A/G, -308 A/G, -857 C/T, -863 A/C) were in association with the outcome of HBV infection in a sample of Iranian population.
3. Methods
3.1. Study Subjects
In this study, 100 patients with chronic hepatitis B virus (HBV group) (mean age of 29.03 ± 5.710 and age range of 18 - 43), 40 spontaneously recovered subjects (SR group) (mean age of 29.72 ± 5.517 and age range of 18 - 42), and 100 healthy controls (C group) (mean age of 30.44 ± 4.539 and age range of 22 - 45) were recruited. The participants were selected among those who referred to the blood transfusion organization clinics in Zahedan, Iran, between July and December 2015. Participants were selected based on the following inclusion criteria: HBV patients were positive for HBsAg and antibodies against anti-HBc and had impaired liver function test so that their transaminases were twofold more than the normal level for at least six months. In HBV group, serological tests (presence of HBsAg by enzyme-linked immunosorbent assay (ELISA)) and clinical findings were compatible with chronic liver disease. The spontaneously recovered group was negative for HBsAg and anti-HBc and positive for antibodies against anti-HBs and anti-HBc. The control group comprised HBsAg, anti-HBs, and anti-HBc negative, healthy volunteers with normal values for alanine transaminase (ALT), without any history of hepatitis B infection. All subjects were in the same geographical area. There was no difference between groups in terms of age, gender, and ethnicity. Subjects with HCV, HEV, HAV, HIV, alcohol consumption, drug abuse, and liver diseases were excluded. All participants were unrelated Iranians.
The study was approved by the institutional ethics committee of Zahedan University of Medical Sciences (grant number: 6809) and carried out in Infectious Diseases and Tropical medicine research center, Zahedan, Iran. Written informed consent was obtained from each participant.
3.2. Analysis of the Serum TNF-α
Three ml of blood was taken in tubes containing EDTA for determining biochemical parameters. The serum level of TNF-α was measured using a sensitive sandwich ELISA technique (EASTBIOPHARM, China), according to the manufacturer’s instructions. Briefly, serially diluted standards and samples were added to the 96-well plates in triplicate. Enzyme-labeling reagents (50 mL) were then added to each well, and plates were incubated for 30 minutes at 37°C. Subsequently, chromogenic substrates A and B (50 mL each) were added and plates were incubated in the dark at 37°C for 10 minutes to develop signals. The reaction was quenched by addition of a stopping buffer. Plates were read on a microplate reader to record the optical density values at 450 nm. A standard curve was used to determine the sample values using linear regression.
3.3. DNA Extraction
Genomic DNA was extracted from the peripheral blood leukocytes using the salting-out method. Briefly, 500 µL blood was transferred to 1.5-mL microfuge tubes, and 1 mL cell lysis buffer (10 mM Tris-HCl, 11% w/v sucrose, 5 mM MgCl2, and 11% v/v Triton X-100) was added. The solutions were gently mixed and centrifuged for 2 minutes at 6000 rpm at room temperature. Then, the supernatant was discarded. The procedure was repeated twice. Next, 400 µL buffer II (10 mM Tris-HCl, 10 mM EDTA, and 10 mM sodium citrate) and 50 µL 10% SDS were added, and the mixture incubated for 5 minutes at room temperature. Then, 100 µL saturated NaCl and 600 µL chloroform were added with gentle mixing, and the mixture was centrifuged for 2 minutes at 6000 rpm. The supernatant was transferred to a new microfuge tube, where 800 µL cold isopropanol was added, followed by gentle mixing and centrifugation for 1 minute at 12,000 rpm for 2 minutes at 4°C. The supernatant was discarded and 800 µL cold 70% ethanol was added. The suspension was gently mixed and centrifuged for 1 minute at 12,000 rpm at 4°C. Pellets were subsequently dried before dissolving in 100 µL distilled water. These samples were stored at -80°C until they were used for the study.
3.4. Polymorphism Genotyping
The TNF-α polymorphisms (-238 A/G, -308 A/G, -857 C/T, and -863 A/C) were detected by polymerase chain reaction-restriction fragment-length polymorphism (PCR-RFLP). The primers were designed according to the data achieved from the NCBI data bank (http:www.ncbi.nlm.nih.gov) for identification of SNPs and listed in Table 1. The restriction enzymes and the fragment length after digestion are shown in Table 1. Every reaction contained 1 μL of each primer, 100 ng of template DNA, and 10 μL of 2X Prime Taq Premix (Genet Bio, Korea) and 7 μL of ddH2O in a 20 μL of total reaction volume.
| Polymorphism | Primers | Annealing Temperature | Restriction Enzyme | Allele Phenotype, bp |
|---|---|---|---|---|
| -238 A/G | F: AGAAGACCCCCCTCGGAACC | 58.5°C | Msp I | A: 152; G: 132 + 20 |
| R: ATCTGGAGGAAGCGGTAGTG | ||||
| -308 A/G | F: ATCTGGAGGAAGCGGTAGTG | 59°C | NcoI | A: 222; G: 206 + 16 |
| R: AATAGGTTTTGAGGGCCATG | ||||
| -857C/T | F: AAGTCGAGTATGGGGACCCCCCGTTAA | 63°C | HincII | C: 108 + 25; T: 133 |
| R: CCCCAGTGTGTGGCCATATCTTCTT | ||||
| -863 A/C | F:ATGTAGCGGCTCTGAGGAATGGGTTACA | 52°C | StyI | A: 132; C: 108 + 24 |
| R: CTACATGGCCCTGTCTTCGCCAAG |
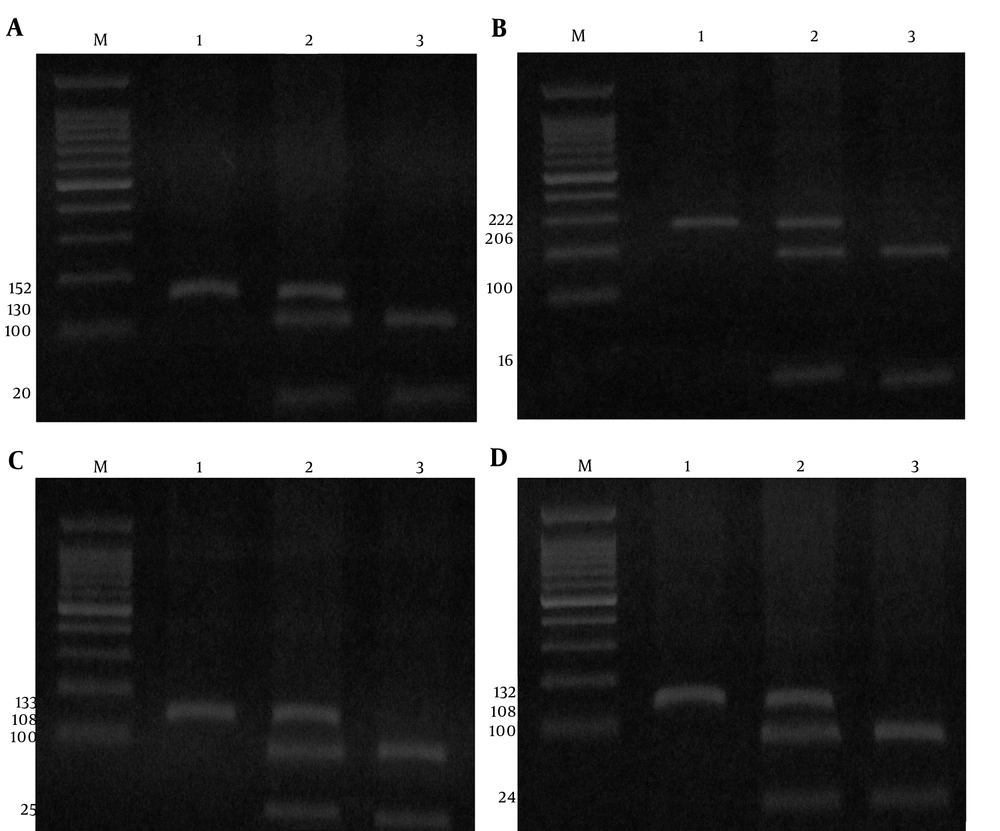
The PCR conditions were as follows: initial denaturation at 95°C for 5 minutes followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 58.5°C for -238 A/G, 59°C for -308 A/G, 63°C for -857 C/T, and 52°C for -863 A/C polymorphisms of the TNF-α for 30 seconds and extension at 72°C for 30 seconds, followed by a final extension step at 72°C for 5 minutes. Finally, the PCR products digested by the restriction enzymes (Fermentas, Vilnius, and Lithuania) and digested products were resolved by electrophoresis in 4 % agarose gel and stained with ethidium bromide (Figure 1A, B, C, and D).
(A), The digestion pattern of Msp I restriction enzyme on 4% agarose gel at -238 A/G SNP; M marker 100bp, 1 genotype AA, 2 genotype AG, 3 genotype GG; (B), The digestion pattern of NcoI restriction enzyme on 4% agarose gel at -308 A/G SNP; M marker 100bp,1 genotype AA, 2 genotype AG, 3 genotype GG; (C), The digestion pattern of HincII restriction enzyme on 4% agarose gel at -857 C/T SNP, M marker 100 bp,1 genotype TT, 2 genotype CT, 3 genotype CC; (D), The digestion pattern of StyI restriction enzyme on 4% agarose gel at -863 A/C SNP; M marker 100bp,1 genotype AA, 2 genotype AC, 3 genotype CC.
3.5. Statistical Analysis
Data were reported as mean values ± SD for parametric variables and as percentages for non-parametric values. The statistical analyses were performed using SPSS Statistical package, version 17.0 for Windows (SPSS. Inc., USA). Pearson’s χ2 test (for categorical variables) and Kruskal-Wallis H test (for continuous variables including age) were used to examine differences among the three groups in the distributions of demographic characteristics and genotype frequencies. The frequency of the genotypes in patients and control groups was estimated by direct gene counting. The risk of HBV infection associated with SNPs was estimated using odds ratios (ORs) and 95% confidence intervals (95% CIs) calculated by a multivariate logistic regression model after adjustment for sex, age, and ethnicities. All P values were two tailed. A P value of less than 0.05 was considered significant at 95% confidence interval (CI).
4. Results
Clinical characteristics of study population are listed in Table 2. Study subjects were divided into three groups: patients with chronic HBV infection (female/male: 43.0/57.0), patients who spontaneously recovered from infection (female/male: 37.5/62.5), and healthy people (female/male: 49.0/51.0). The gender ratio (female/male) was not significantly different between the 3 groups (P = 0.427). The mean age in the study groups was not statistically different (P = 0.125). The prevalence of various ethnicities was also not different between HBV, SR, and C groups (P = 0.292).
| Parameters | C | SR | HBV | P |
|---|---|---|---|---|
| Age, y | 30.44 ± 4.539 | 29.72 ± 5.517 | 29.03 ± 5.710 | 0.125 |
| Sex | ||||
| Male | 51 (51.0) | 25 (62.5) | 57 (57.0) | 0.427 |
| Female | 49 (49.0) | 15 (37.5) | 43 (43.0) | |
| Ethnicity | ||||
| Sistani | 46 (46.0) | 18 (45.0) | 41 (41.0) | 0.292 |
| Baluch | 18 (18.0) | 13 (32.5) | 22 (22.0) | |
| Others | 36 (36.0) | 9 (22.5) | 37 (37.0) |
aValues are expressed as No. (%).
4.1. Genotype Analysis
Genotype results at positions -238 A/G, -308 A/G, -857 C/T, and -863 A/C in the TNF-α gene are listed in Tables 3 and 4. The distribution of the polymorphisms was in Hardy-Weinberg equilibrium (P > 0.05). For -308 A/G, -857 C/T, and -863 A/C SNPs, we found that compared to the healthy group (C + SR), chronic HBV patients have significantly higher G, C, and A alleles and GG, CC/CT, and AA/AC genotypes frequency, respectively. This study showed that the existence of -308 G, -857 C, and -863 A alleles significantly increased the risk of chronic HBV infection compared to the other alleles. Also, these results showed that there is not any relationship between -308 G, -857 C, and -863 A alleles and elimination of HBV. For -238 A/G polymorphism, no significant differences were found in allele or genotype frequencies between the groups.
| TNF-α Polymorphisms | C (%) | SR (%) | P Value | Odds Ratio | HBV (%) | P Value | Odds Ratio |
|---|---|---|---|---|---|---|---|
| -238 A/G | |||||||
| AA | 3 (3.0) | 1 (2.5) | Ref = 1 | - | 2 (2.0) | Ref = 1 | - |
| AG | 5 (5.0) | 2 (5.0) | 0.898 | 1.200 (0.073 - 19.631) | 3 (3.0) | 0.928 | 0.900 (0.091 - 8.899) |
| GG | 92 (92.0) | 37 (92.5) | 0.873 | 1.207 (0.122 - 11.975) | 95 (95.0) | 0.636 | 1.549 (0.253 - 9.484) |
| GG + AG | 97 (97.0) | 39 (97.5) | 0.873 | 1.206 (0.122 - 11.953) | 98 (98.0) | 0.653 | 1.515 (0.248 - 9.270) |
| A | 11 (5.5) | 4 (5.0) | Ref = 1 | - | 6 (3.0) | Ref = 1 | - |
| G | 189 (94.5) | 76 (95.5) | 0.867 | 1.106 (0.342 - 3.581) | 194 (97.0) | 0.222 | 1.882 (0.682 - 5.191) |
| -308 A/G | |||||||
| AA | 7 (7.0) | 5 (12.5) | Ref = 1 | - | 2 (2.0) | Ref = 1 | - |
| AG | 29 (29.0) | 10(25.0) | 0.292 | 0.483 (0.125 - 1.870) | 6 (6.0) | 0.725 | 0.724 (0.120 - 4.383) |
| GG | 64 (64.0) | 25 (62.5) | 0.339 | 0.547 (0.159 - 1.885) | 92 (92.0) | 0.048 | 5.031 (1.012 - 25.008) |
| GG + AG | 93 (93.0) | 35 (87.5) | 0.300 | 0.527 (0.157 - 1.770) | 98 (98.0) | 0.109 | 3.688 (0.747 - 18.211) |
| A | 43 (21.5) | 20 (25.0) | Ref = 1 | - | 10 (5.0) | Ref = 1 | - |
| G | 157 (78.5) | 60 (75.0) | 0.527 | 0.822 (0.447 - 1.509) | 190 (95.0) | 0.000 | 5.204 (2.533 - 10.689) |
| -857 C/T | |||||||
| CC | 72 (72.0) | 25(62.5) | 0.349 | 0.663 (0.281 - 1.566) | 80 (80.0) | 0.016 | 2.917 (1.217 - 6.992) |
| CT | 7 (7.0) | 4 (10.0) | 0.905 | 1.091 (0.261 - 4.553) | 12 (12.0) | 0.017 | 4.500 (1.305 - 15.515) |
| TT | 21 (21.0) | 11 (27.5) | Ref = 1 | - | 8 (8.0) | Ref = 1 | - |
| CC + CT | 79 (79.0) | 29 (72.5) | 0.409 | 0.701 (0.301 - 1.631) | 92 (92.0) | 0.012 | 3.057 (1.283 - 7.282) |
| C | 151 (75.5) | 54 (67.5) | 0.173 | 0.674 (0.382 - 1.189) | 172 (86.0) | 0.008 | 1.993 (1.193 - 3.330) |
| T | 49 (24.5) | 26 (32.5) | Ref = 1 | - | 28 (14.0) | Ref = 1 | - |
| -863 A/C | |||||||
| AA | 5 (5.0) | 1 (2.5) | 0.499 | 0.469 (0.052 - 4.216) | 12 (12.0) | 0.010 | 3.955 (1.391 - 11.243) |
| AC | 34 (34.0) | 13 (32.5) | 0.787 | 0.897 (0.408 - 1.970) | 44 (44.0) | 0.028 | 1.851 (1.070 - 3.203) |
| CC | 61 (61.0) | 26 (65.0) | Ref = 1 | - | 44 (44.0) | Ref = 1 | - |
| AA + AC | 39 (39.0) | 14 (35.0) | 0.660 | 0.842 (0.392 - 1.808) | 56 (56.0) | 0.006 | 2.089 (1.240 - 3.521) |
| A | 44 (22.0) | 15 (18.8) | 0.547 | 0.818 (0.426 - 1.573) | 68 (34.0) | 0.008 | 1.826 (1.171 - 2.849) |
| C | 156 (78.0) | 65 (81.2) | Ref = 1 | - | 132 (66.0) | Ref = 1 | - |
| TNF-α Polymorphisms | HBV (%) | Healthy(C + SR) (%) | P Value | Odds Ratio |
|---|---|---|---|---|
| -238 A/G | ||||
| AA | 2 (2.0) | 4 (2.9) | Ref = 1 | - |
| AG | 3 (3.0) | 7 (5.0) | 0.889 | 0.857 (0.098 - 7.510) |
| GG | 95 (95.0) | 129 (92.1) | 0.659 | 1.473 (0.264 - 8.208) |
| GG + AG | 98 (98.0) | 136 (97.1) | 0.677 | 1.441 (0.259 - 8.025) |
| A | 6 (3.0) | 15 (5.4) | Ref = 1 | - |
| G | 194 (97.0) | 265 (94.6) | 0.219 | 1.830 (0.697-4.802) |
| -308 A/G | ||||
| AA | 2 (2.0) | 12 (8.6) | Ref = 1 | - |
| AG | 6 (6.0) | 39 (27.9) | 0.928 | 0.923 (0.164 - 5.187) |
| GG | 92 (92.0) | 89 (63.6) | 0.019 | 6.202 (1.350 - 28.502) |
| GG + AG | 98 (98.0) | 128 (91.4) | 0.049 | 4.594 (1.005 - 21.001) |
| A | 10 (5.0) | 63 (22.5) | Ref = 1 | - |
| G | 190 (95.0) | 217 (77.5) | 0.000 | 5.516 (2.753 - 11.053) |
| -857 C/T | ||||
| CC | 80 (80.0) | 97 (69.3) | 0.005 | 3.299 (1.439 - 7.561) |
| CT | 12 (12.0) | 11 (7.9) | 0.010 | 4.364 (1.414 - 13.465) |
| TT | 8 (8.0) | 32 (22.9) | Ref = 1 | - |
| CC + CT | 92 (92.0) | 108 (77.1) | 0.004 | 3.407 (1.496 - 7.761) |
| C | 172 (86.0) | 205 (73.2) | 0.001 | 2.247 (1.392 - 3.628) |
| T | 28 (14.0) | 75 (26.8) | Ref = 1 | - |
| -863 A/C | ||||
| AA | 12 (12.0) | 6 (4.3) | 0.010 | 3.955 (1.391 - 11.243) |
| AC | 44 (44.0) | 47 (33.6) | 0.028 | 1.851 (1.070 - 3.203) |
| CC | 44 (44.0) | 87 (62.1) | Ref = 1 | - |
| AA + AC | 56 (56.0) | 53 (37.9) | 0.006 | 2.089 (1.240 - 3.521) |
| A | 68 (34.0) | 59 (21.1) | 0.002 | 1.930 (1.281 - 2.908) |
| C | 132 (66.0) | 221 (78.9) | Ref = 1 | - |
We conducted the -238 A/G, -308 A/G, -857 C/T, and -863 A/C haplotype association analyses and found 5 genotypes. As shown in Tables 5 and 6, the distribution frequency of haplotypes showed a significant association with chronic HBV infection compared to the C and SR groups. In our study, GGCA, GGCC, and GGTA haplotypes had a higher frequency in HBV patients than C and SR groups. Also, -238 A/G, -308 A/G, -857 C/T, and -863 A/C polymorphisms indicated enrichment of GGCA haplotype among patients than healthy controls (C + SR) (48.0% vs. 30.7, P = 0.005). It means that this haplotype might relate to the natural history of the infection. Linkage disequilibrium (LD) analysis did not show any significant statistical relationship between any of the 4 positions.
| Haplotypes | C Group | SR Group | P Value | Odds Ratio | HBV Group | P Value | Odds Ratio |
|---|---|---|---|---|---|---|---|
| GGCC | 27 (27.0) | 10 (25.0) | 0.523 | 1.605 (0.376 - 6.842) | 33 (33.0) | 0.010 | 7.944 (1.648 - 38.308) |
| GGCA | 30 (30.0) | 13 (32.5) | 0.382 | 1.878 (0.457 - 7.722) | 48 (48.0) | 0.003 | 10.400 (2.192 - 49.344) |
| AGCC | 16 (16.0) | 7 (17.5) | 0.415 | 1.896 (0.407 - 8.824) | 4 (4.0) | 0.607 | 1.625 (0.256 - 10.318) |
| GACC | 13 (13.0) | 3 (7.5) | Ref = 1 | - | 2 (2.0) | Ref = 1 | - |
| GGTA | 14 (14.0) | 7 (17.5) | 0.328 | 2.167 (0.460 - 10.197) | 13 (13.0) | 0.035 | 6.036 (1.137 - 32.036) |
| Total | 100 (100.0) | 40 (100.0) | 100 (100.0) |
aValues are expressed as No. (%).
| Haplotypes | Healthy (C + SR) | HBV Group | P Value | Odds Ratio |
|---|---|---|---|---|
| GGCC | 37 (26.4) | 33 (33.0) | 0.013 | 7.135 (1.525 - 33.385) |
| GGCA | 43 (30.7) | 48 (48.0) | 0.005 | 8.930 (1.941 - 41.097) |
| AGCC | 23 (16.4) | 4 (4.0) | 0.721 | 1.391 (0.227 - 8.530) |
| GACC | 16 (11.4) | 2 (2.0) | Ref = 1 | - |
| GGTA | 21 (15.0) | 13 (13.0) | 0.054 | 4.952 (0.976 - 25.140) |
| Total | 140 (100.0) | 100 (100.0) |
aValues are expressed as No. (%).
4.2. Analysis of Serum TNF-α Level
TNF-α levels in HBV patients, SR subjects, and C group were 1.41 ± 0.062 (pg/mL), 1.99 ± 0.159 (pg/mL), and 2.03 ± 0.193 (pg/mL), respectively. We found a significant difference in the serum levels of TNF-α between the groups (P < 0.05). The serum levels of TNF-α significantly decreased in the chronic HBV patients compared to the healthy groups (C + SR) (P < 0.05). Statistical analysis indicated positive correlations between TNF-α genotypes and serum levels of the TNF-α in HBV patients (Table 7). Chronic HBV patients with -308 GG/AG, -857 CC, and -863 AA genotypes had lower levels of TNF-α compared to those with -308 AA (P = 0.000/0.001), -857 TT (P = 0.009), and -863CC (P = 0.041) genotypes, respectively. The results showed that the risk of HBV infection significantly increased in subjects with at least one of -308G, -857C, and -863A alleles that were effective in reducing the levels of TNF-α, compared to the other alleles (Table 8).
| TNF-α Genotype | HBV | HBV TNF-α, pg/mL | Healthy (C + SR) | Healthy (C + SR) TNF-α, pg/mL |
|---|---|---|---|---|
| -238 A/G | ||||
| AA | 2 (2.0) | 1.43 ± 0.007 | 4 (2.9) | 1.93 ± 0.050 |
| AG | 3 (3.0) | 1.43 ± 0.049 | 7 (5.0) | 1.98 ± 0.158 |
| GG | 95 (95.0) | 1.41 ± 0.063 | 129 (92.1) | 2.02 ± 0.188 |
| P | 0.837 | 1.000 | ||
| F | 0.179 | 0.175 | ||
| -308 A/G | ||||
| AA | 2 (2.0) | 1.73 ± 0.134 | 12 (8.6) | 1.94 ± 0.139 |
| AG | 6 (6.0) | 1.46 ± 0.198a | 39 (27.9) | 2.00 ± 0.156 |
| GG | 92 (92.0) | 1.40 ± 0.081b | 89 (63.6) | 2.04 ± 0.198 |
| P | 0.000 | 0.974 | ||
| F | 13.138 | 0.446 | ||
| -857 C/T | ||||
| CC | 80 (80.0) | 1.40 ± 0.105c | 97 (69.3) | 2.00 ± 0.166 |
| CT | 12 (12.0) | 1.46 ± 0.091 | 11 (7.9) | 1.98 ± 0.153 |
| TT | 8 (8.0) | 1.52 ± 0.153 | 32 (22.9) | 2.08 ± 0.233 |
| P | 0.004 | 0.356 | ||
| F | 5.891 | 1.105 | ||
| -863 A/C | ||||
| AA | 12 (12.0) | 1.37 ± 0.149d | 6 (4.3) | 1.95 ± 0.086 |
| AC | 44 (44.0) | 1.40 ± 0.082 | 47 (33.6) | 1.98 ± 0.160 |
| CC | 44 (44.0) | 1.44 ± 0.067 | 87 (62.1) | 2.04 ± 0.198 |
| P | 0.023 | 0.944 | ||
| F | 3.914 | 0.520 |
aP = 0.001, Compared to genotype AA.
bP = 0, Compared to genotype AA.
cP = 0.009, Compared to genotype TT.
dP = 0.041, Compared to genotype CC.
| Haplotypes | HBVN | HBVTNF-α, pg/mL | Healthy (C + SR) | Healthy (C + SR) TNF-α, pg/mL |
|---|---|---|---|---|
| GGCC | 33 (33.0) | 1.43 ± 0.100 | 37 (26.4) | 2.00 ± 0.164 |
| GGCA | 48 (48.0) | 1.35 ± 0.128 | 43 (30.7) | 1.98 ± 0.155 |
| AGCC | 4 (4.0) | 1.81 ± 0.107 | 23 (16.4) | 2.05 ± 0.183 |
| GACC | 2 (2.0) | 1.52 ± 0.028 | 16 (11.4) | 2.01 ± 0.158 |
| GGTA | 13 (13.0) | 1.45 ± 0.055 | 21 (15.0) | 2.0 ± 0.268 |
| TOTAL | 100 (100.0) | P = 0.984; F = 0.484 | 140 (100.0) | P = 0.164; F = 1.360 |
aValues are expressed as No. (%) or mean ± SD.
5. Discussion
The current study showed a significant association between TNF-α polymorphism and susceptibility to chronic HBV infection, which support some previous studies in other countries and emphasizes the critical role of TNF-α in chronic HBV infection. Our results showed that -308 GG, -857 CC/CT, and -863AA/AC genotypes were associated with the susceptibility to the HBV infection, and -308 G, -857 C, and -863 A alleles were clearly related to the disease. In addition, the GGCA haplotype was more frequent in HBV patients, confirming that TNF-α polymorphism can be a risk factor for the infection.
It has been recognized that host genetic background same as virus profile, can affect the outcome of HBV infection (26). A study on twins showed an increased rate of HBV infection in identical twins in comparison with non-identical twins, which indicates the effect of host genetic on the disease (3).
TNF-α, as an important pro-inflammatory cytokine, can also regulate transcription of other immune regulatory proteins, which amplify immune responses against infections (3). In vitro studies have indicated inhibitory effects of TNF-α on the HBV virus replication and increasing HBV mRNA destruction (27). Also, serum level of TNF-α may be associated with the progression of the disease (28). It has been revealed that the amount of TNF-α was significantly different in HBV patients compared to the simultaneously recovered subjects from HBV (29). An in vivo study found that TNF-α reduced infection by destabilizing HBV nucleocapsids and reducing the cccDNA (30).
In this regard, SNPs in promoter region might change the expression of protein at the transcriptional and post-transcriptional levels. Some studies have identified TNF-α promoter SNPs and their association with the susceptibility to HBV infection, but the results are conflicting because of the different inclusion criteria, various genotypes of the virus, and insufficient sample size.
In our study, the results revealed a significantly increased risk of HBV infection because of the presence of the TNF-α -308 G, -857 C, and -863 A alleles in the Iranian population.
The TNF-α gene is linked to a cluster of genes such as lymphotoxin-a (LTA) that relates to the disease progression (31). TNF-α and LTA are homotrimer cytokines. TNF-α can bind to the TNF receptors 1 and 2 on parenchymal cells (such as hepatocytes) and leucocytes and then mediate immune responses against various infectious pathogens in human (32). In addition, TNF-α polymorphisms regulate the RNA polymerase II-binding and allele-specific LTA transcription in the host immune system (33).
The current study did not demonstrate a significant association between -238 A/G polymorphism and HBV infection. However, the expression of TNF-α is affected by SNPs, but we did not find any difference among -238 A/G genotypes with regard to the serum levels of TNF-α. In accordance with our findings, Du et al. (34) and Lu et al. (35) reported an association between the -238 GG genotype and HBV infection. The current study indicated a relative increase in the frequency of -238 GG genotype and G allele in HBV infected patients; but it was not significant. The lack of significance is probably attributed to the complication of the human’s immune response and also may be due to the low sample size and other genetic or environmental factors. By the way, the analysis of this polymorphism in HBV infection has shown conflicting results. It has been determined that the binding capabilities of allele-specific binding elements with trans factors mediated the relationship between polymorphisms and protein expression. TNF-α -238 A/G is located in the regulatory sequences of the promoter named Y-box which can bind to the DNA-binding proteins. Therefore, genetic variation in the Y-box will change the functional activity of the promoter (36).
The results showed that the frequency of -308 GG genotype in the chronic HBV patients was higher than that in the SR and C groups. Also, in HBV patients the amount of TNF-α decreased, which could be explained by the existence of the -308 G allele in a way that -308 G can inhibit the production of the TNF-α and then intensify the disease. Our findings were in agreement with the results reported by Basturk et al. (37), Xu et al. (38), Xia et al. (39), Kim et al. (24), and Zheng et al. (40). They highlighted the key role of -308 A/G polymorphism both in susceptibility to HBV infection and increased serum TNF-α level in HBV patients and emphasized that the presence of G at the position -308 increased the risk of HBV infection. However, our findings were different from the results of some studies (41-43) claiming that the A allele may have a role in HBV aggravation. In addition, Somi et al. (44) found that TNF-α promoter polymorphism -308 was common in Iranian population, but they did not find any association between this SNP and development of chronic HBV infection. Their failure to find an association may be related to the type II error, and larger studies may provide more helpful results. Also, it seems that ethnic differences have an effect on the distribution of cytokine gene polymorphisms. Therefore, variation in the study population could account for the different frequencies in our population and these contradictions might be due to different study populations.
Increased distribution of the -857 (CC + CT) genotypes in the HBV patients might be due to lower transcriptional activities in subjects with the C allele. It was revealed that -857 TT genotype is associated with higher expression of TNF-α and inhibition of HBV infection in subjects spontaneously recovered from HBV (45). It was also noted that, TNF-α -857 C/T polymorphism was located in the binding sequence of the octamerbinding protein (OCT1). -857 C/T SNP might affect the interaction between the OCT1 and binding sequence, leading to a change in the transcriptional function of the gene (46). In the line with our findings, Hohjoh et al. (47) and Zhang et al. (48) reported that -857 C allele is associated with the reduced production of the TNF-α and increased the risk of disease because of the afore mentioned mechanism. Also, it is probable that the combination of transcriptional regulators of HBV to the gene promoter changes with the presence of SNPs, which causes different trans activities.
Our study indicated the -863 A/C polymorphism as a risk factor for chronic HBV infection. We found that the existence of at least one -863 A allele in subjects increased the severity of the infection, which was associated with lower serum TNF-α levels (49). Also, we observed that TNF-α production reduced in subjects with A allele compared to the subjects with the C allele among the HBV patients. In agreement with our findings, in vivo studies with the hepatoblastoma cell line indicated that -863 A allele is associated with the reduction of TNF-α receptor expression (49, 50). It should be noted that -863 A allele can bind to the p50/p65 complex of the downstream nuclear factor-kB (NF-κB) transcription factor, and C allele can bind to both p50/p50 and p50/p65. This different affinity leads to the change in the downstream NF-κB activation (51). Therefore, -863A/C various genotypes might be responsible for the different TNF-α serum levels and susceptibility to the HBV infection. Conversely, Xu et al. (38), and Gao et al. (52) did not find statistically significant difference in TNF-α -863 polymorphism between HBV patients and healthy controls.
The haplotype (-238 A/G, -308 A/G, -857 C/T, and -863 A/C) analysis revealed that the GGCA haplotype with low production of TNF-α was a potential risk factor for HBV infection. Interestingly, haplotype analysis showed a strong association between TNF-α and HBV, and emphasized that the susceptibility to disease may be due to the influence of TNF-α polymorphisms on the promoter region of the gene.
Consequently, we can obtain a reasonable evidence of the relationship between chronic hepatitis B and TNF-α polymorphisms. Our results demonstrate an increased risk for chronic HBV infection with TNF-α -308 GG, -857 CC, and -863 AA genotypes. Also, the carriers of -238 G, -308 G, -857 C, and -863 A alleles with lower amount of TNF-α have a serious risk of infection compared to the subjects who do not have these alleles. The GGCA haplotype is a risk factor for disease progression. The identification of appropriate diagnostic biomarkers would develop the best diagnostic approach and clinical management for HBV.
Some disagreements with the current study indicate the possible impact of genetic background of different populations. On the other hand, several factors, such as sample size, patients’ selection, different cultural backgrounds, epidemiological and geographical factors, study conditions (such as number and characteristics of the subjects and HBV genotype variations) and different gene-gene interactions also can cause importing results. Additional studies with regard to the abovementioned limitations should provide additional understanding of TNF-α gene variant in chronic HBV infection and provide good predictors of disease and therapeutic tools for subjects who are at risk.