1. Context
Hepatitis C virus (HCV) infection is still one of the major causes of mortality and morbidity worldwide (1, 2). The world health organization (WHO) has estimated 3% prevalence of HCV equating approximately to 180 million individuals globally (3-5). The prevalence of HCV infection in Iran is estimated to be less than 0.5% (6, 7). It is well established that hepatitis C contributes to the increasing risk of fatal-related diseases including cirrhosis and hepatocellular carcinoma (HCC) (8, 9). In spite of numerous progressions in hepatitis C treatment, the high prevalence of hepatitis C in developing countries is still a major concern (10).
The only one open reading frame (ORF) of HCV comprises about 9024 base pairs and encodes a polyprotein of about 3000 amino acids. Up to now, HCV is classified into seven different genotypes and more than 67 subtypes according to the genetic variability and viral sequences (11). The linkage of geographic distribution of HCV genotypes in different populations and specific risk groups to genetic diversity of HCV is obvious (12). Genotype of HCV is a considerable factor with clinical and epidemiological importance because it determines the rate of response to the HCV therapy, and may help trace the source of infection and clarify the possible modes of transmission (13). In terms of response to the HCV treatment protocol with Pegylated-Interferon and Ribavirin (PegIFN/RBV), the HCV genotype 1 (HCV-1) and HCV-4 infections are more difficult-to-treat than HCV-2 and HCV-3 infections (10, 14, 15). However, the development of new pangenotypic HCV treatments in recent years has contributed to the HCV elimination in the world (13).
The Eastern Mediterranean regional office (EMRO) is one of the six regional divisions of the WHO around the world, which serves 22 countries and territories in the Middle East, the North Africa, the Horn of Africa, and Central Asia with a total population of 605 million people. The frequency of hepatitis C infection, as estimated by the WHO, has revealed that at least 23 million individuals in the EMRO countries are infected with HCV (16). The genotype distribution of HCV through EMRO countries is interestingly heterogeneous. The distribution of HCV genotypes in EMRO countries has two main patterns: Arab countries (except Jordan) with HCV-4 as the predominant isolate and non-Arab countries with predominance of HCV genotypes other than HCV-4 such as Iran by dominance of HCV-1 and Afghanistan and Pakistan by dominance of HCV-3 (17, 18). Distribution of HCV genotypes in Iran is different from most of the other neighboring and Middle Eastern countries, as the most frequent HCV genotype in Iran is HCV-1a, followed by HCV-3a and -1b, respectively (19). Interestingly, in Turkey, Azerbaijan and Russia, as neighboring countries in North of Iran, the most prevalent genotype is HCV-1b (20), in Afghanistan and Pakistan in East of Iran, the most prevalent genotype is HCV-3 (21), and in most of the neighboring Arab countries in West and South of Iran including Iraq and Saudi Arabia, the most prevalent genotype is HCV-4 (18, 22, 23). It is thought that the genetic diversity patterns of HCV in Iran are similar to the pattern observed in North America and somewhat Western Europe (24). The controversial issue regarding the source of HCV in Iran in comparison with surrounding countries may be elucidated by phylogenetic analysis.
There is heterogeneity in the regions sequenced along the HCV genome, such as Core, NS5B, HVR-1, E2, and a segment of the NS5A gene associated with interferon sensitivity. Guidelines propose to use either the full genome, Core/E1 or NS5B sequences of HCV for classification of genotypes/subtypes (25). Furthermore, genotyping of HCV by nucleotide sequence analysis of NS5B is an effective procedure that allows discrimination of HCV subtypes properly. Moreover, NS5B is an appropriate gene region to study the molecular epidemiology of HCV (25).
Phylogenetic is a study for inferring or estimating evolutionary relationships among individuals or groups of organisms e.g. species or populations (26). In the area of virology, phylogenetic trees contain a lot of information about the inferred evolutionary relationships between a set of viruses and it can be a potential widely used molecular tool to study rapidly-evolving RNA viruses such as HCV.
The main objective of the current study was to investigate the genetic relationship among all HCV-1a and -1b sequences derived from Iran, EMRO, Middle Eastern, and some European and North American countries by applying phylogenetic analysis, and understanding of the source of spread of HCV-1a and -1b in Iran.
2. Evidence Acquisition
2.1. Search Strategy
An electronic systematic search of available systematic reviews was conducted on all literature to find relevant studies reporting molecular prevalence and evaluation of HCV-1a and -1b in different Iranian patient groups, and also all studies about HCV-1 molecular epidemiology in EMRO and Middle Eastern countries. To compare the obtained findings, we also extended the search on some European and North American countries.
The search was performed on all peer-reviewed journals indexed in PubMed, Scopus, and Web of Science databases. The literature review was carried out using the following key terms: “hepatitis C virus”, “HCV”, “genotype”, “genotype 1a and 1b”, “molecular sequence data”, “sequence analysis”, “phylogeography”, “phylogenetic analysis”, and “Iran”. In addition to aforementioned search terms, the names of twenty-two EMRO countries were added to our search as follows: Afghanistan, Bahrain, Djibouti, Egypt, Iran, Iraq, Jordan, Kuwait, Lebanon, Libya, Morocco, Oman, Pakistan, Palestine, Qatar, Saudi Arabia, Somalia, Sudan, Syria, Tunisia, United Arab Emirates, and Yemen. Moreover, Middle Eastern countries that were not in EMRO including Cyprus and Turkey were added in the search. Furthermore, Azerbaijan was added in the search, as a neighboring country of Iran with wide commuting between the countries. Also, some European and North American countries including France, Germany, Italy, Netherlands, Spain, UK, USA and Canada, were added to the search strategy.
In addition, to find appropriate sequences from unpublished studies which were registered in the GenBank database, we searched into GenBank using the names of aforementioned EMRO and Middle Eastern countries in addition to “Hepatitis C virus” and “NS5B”. After all searches were completed, the NS5B sequences of all HCV-1a and -1b were selected and FASTA format of the sequences were extracted from the GenBank database.
2.2. Selection of Studies
All published and unpublished studies with proper data were surveyed according to the following criteria: 1) all molecular studies in English among different patient groups with HCV-1a and -1b enrolled from Iran, EMRO, Middle Eastern, and some aforementioned European and North American countries, 2) molecular studies that reported nucleotide sequence accession numbers based on NS5B gene sequences which were registered in the GenBank database, 3) gene sequences with 243 base pair (bp) coverage which were between nucleotides 8319 - 8561 for HCV-1a (the coverage obtained after full alignment along with HCV-1a reference with accession number M62321), and 232 bp coverage between nucleotides 8315 - 8546 for HCV-1b (the coverage obtained after full alignment along with HCV-1b reference with accession number U84014). The mentioned frames were obtained after alignment and trimming of the included nucleotide sequences and exclusion of the sequences with less than 200 bp lengths. Generally, the short nucleotide sequences or the nucleotide sequences which were not in the proper coverage were removed from the MEGA file.
The exclusion criteria were as follows: 1) studies with possible errors and confusing data, 2) studies that used HCV genomic regions other than NS5B, 3) the HCV-2 to -6 isolates and also, HCV-1 other than HCV-1a and -1b isolates.
To confirm the genotype/subtype of the selected isolates, we used NCBI Viral Genotyping tool (http://www.ncbi.nlm.nih.gov/projects/genotyping/ formpage.cgi) and HCV geno2pheno (http://www.geno2pheno.org). For the eligibility criteria, all favorable articles obtained through the search strategy were independently reviewed by three authors (KH-H, H-SH and A-NJTSH). If there was any discrepancy between authors, it was resolved by consulting the supervisor of the study (SMA).
2.3. Data Extraction and Quality Assessment
The reviewing and screening processes in this study were based on the PRISMA guidelines for reporting systematic reviews (27). We independently screened the title, abstract, and full-text of papers identified through the database searches. After full-text screening, the following data were extracted from each study: first author’s name, publication year, country of origin, date of study, type of patient groups, and GenBank accession numbers. All extracted data were systematically double checked by authors independently to avoid any errors. The quality of the included studies was assessed using a modified STROBE checklist (28).
2.4. Sequence Collection and Phylogenetic Analyses
In this study, the evolutionary relationships of isolates were inferred using the Neighbor-Joining method and Kimura 2-Parameter model. The percentage of replicate trees which were associated with clusters was estimated using bootstrap test (500 replicates). The results in this study were based on the clusters identified using the neighbor-joining phylogeny according to bootstrap test with a cut-off value 50% for defining the clusters (values > 50% have been shown next to the branches). The trees were drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree (29).
Pair-wise and multiple alignments of the HCV NS5B sequences were performed using multiple sequence alignment-MUSCLE (Multiple Sequence Comparison by Log-Expectation) through molecular evolutionary genetics analysis software version 7.0 (MEGA 7.0) (30-32). To find the coverage and to edit the nucleotide sequences, the downloaded sequences were transferred to CLC software (CLC Main Workbench 5). Following the full alignment of the sequences and manually trimming, the phylogenetic trees of suitable sequences were constructed by MEGA 7.0 software. Initially, the trees were drawn with traditional-rectangular branch style. Due to large number of data, it would be obscure to show the unclear long traditional tree style. Thus, we decided to convert the trees to circular style for better understanding.
3. Results
3.1. Study Screening and Selection
In the present study, HCV NS5B nucleotide sequences from 30 various studies that had been totally conducted on more than 3000 subjects were collected. We studied all available nucleotide sequences reported from Iran and other EMRO and Middle Eastern countries. Out of seven studies from Iran, four studies were published in 2004 - 2014 (33-36) and three were unpublished. Furthermore, 15 published and unpublished studies from EMRO and Middle Eastern countries including Afghanistan (37), Azerbaijan, Cyprus (38, 39), Egypt, Morocco (40), Pakistan (41), and Tunisia (42, 43) were collected. Furthermore, other studies from the US and European countries were investigated, randomly (44-49).
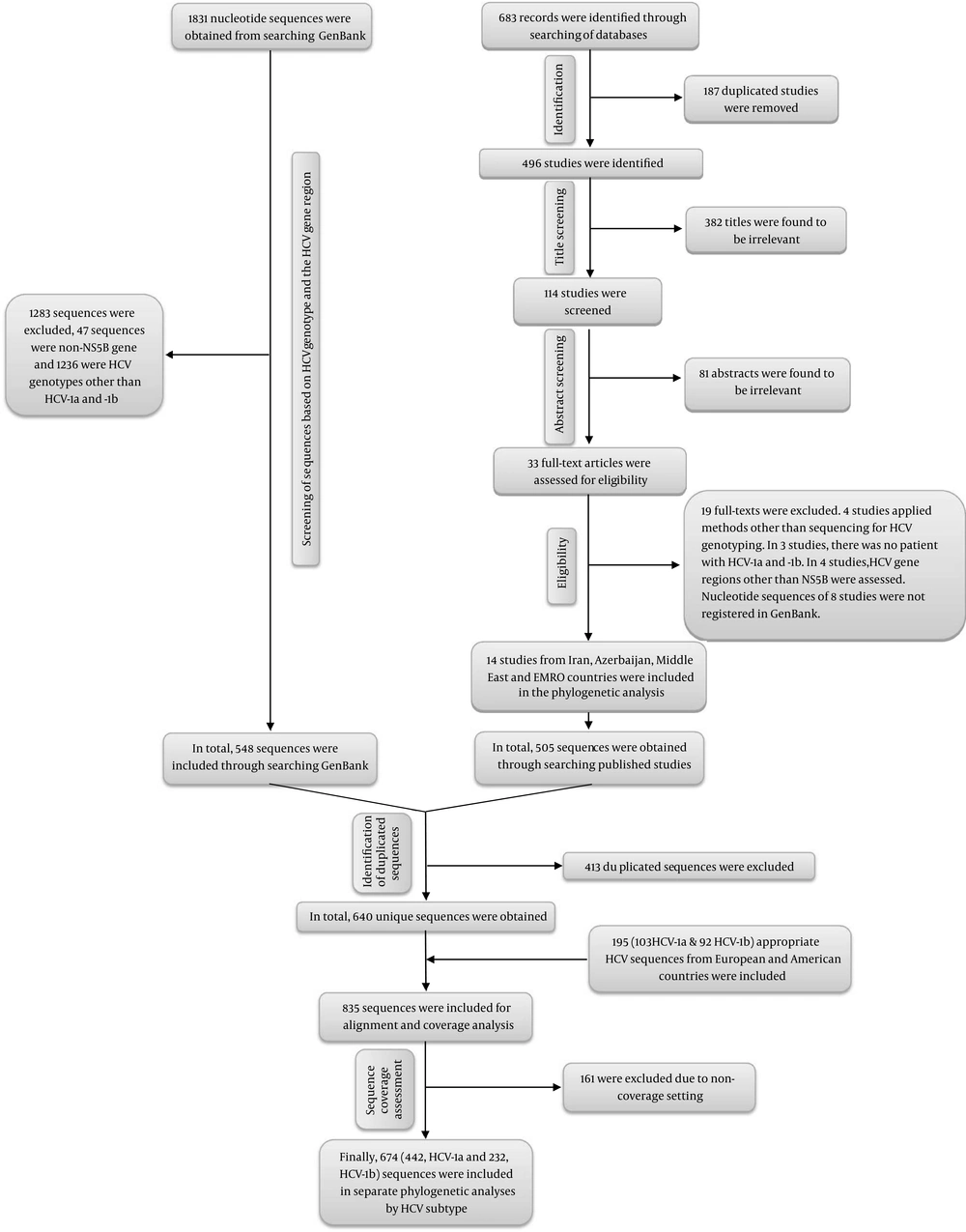
Based on Figure 1, 683 published papers were identified via database searching. After removal of 187 duplicates, 382 irrelevant titles, and 81 papers with irrelevant abstracts, finally, 14 studies were eligible to be assessed in phylogenetic analyses (33-43, 50-52). In total, 505 sequences of HCV-1a and -1b were obtained from these 14 studies. Moreover, after searching the GenBank database to find unpublished studies with registered sequences, 1831 sequences were obtained. After removal of 1283 sequences (including 47 sequences because of non-NS5B gene sequences and 1236 sequences because of HCV genotypes other than HCV-1a and -1b), a total of 548 sequences were obtained. Therefore, a total of 1053 sequences from both published and unpublished studies were collected. After removal of 413 duplicated sequences, 640 sequences were obtained. Furthermore, 195 different sequences from Italy, France, Netherlands, Spain, the United Kingdom, and the United States were randomly included (44-49). After appropriate sequence collection, all of the 835 sequences were transferred into MEGA software for full alignment. Finally, 161 sequences were removed because they were not in the coverage setting; thus, a total of 674 sequences were used in the phylogenetic analyses including 442 sequences for HCV-1a and 232 sequences for HCV-1b.
3.2. Characteristics of the Included Studies
The characteristics of published studies are presented in Table 1. Although 8 included studies were unpublished (from Iran, Azerbaijan, Egypt, Pakistan, and Tunisia), the nucleotide sequences of these studies had been registered in the GenBank database. The characteristics of unpublished studies with registered sequences in the GenBank database are shown in Table 2.
| Publication Year | Country | Sample Size, n | Age, Min - Max | Male, % | Subtype 1a, No. (%) | Subtype 1b, No. (%) | Patient Group | Ref. | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2013 | Afghanistan | 71 | 23-39 | 100 | 25 (35.2) | 2 (2.8) | IDU | (37) |
| 2 | 2009 | Cyprus | 104 | 18 to > 60 | 50 | 9 (8.6) | 38 (36.5) | NA | (38) |
| 3 | 2010 | Cyprus | 40 | 25 - 47 | 85 | 0 | 4 (10) | IDU | (39) |
| 4 | 2004 | Iran | 158 | 5 - 76 | 76 | 59 (37) | 10 (6.3) | IDU, blood or blood product recipient, hemodialysis, NA | (34) |
| 5 | 2012 | Iran | 83 | 19 - 65 | 98 | 35 (42) | 0 | IDU | (35) |
| 6 | 2013 | Iran | 130 | 11 - 63 | 51 | 69 (53) | 19 (14.6) | Thalassemia | (33) |
| 7 | 2014 | Iran | 142 | 22 - 82 | 85 | 71 (50) | 20 (14) | NA | (36) |
| 8 | 2012 | Morocco | 141 | 39 - 80 | 45 | 1 (0.7) | 106 (75) | NA | (40) |
| 9 | 2009 | Pakistan | 189 | 46 - 66 | 66 | 3 (1.5) | 2 (0.8) | NA | (51) |
| 10 | 2013 | Pakistan | 1537 | 31 - 53 | 43 | 53 (3.5) | 12 (0.8) | NA | (41) |
| 11 | 2004 | Tunisia | 32 | 14 - 76 | 81 | 10 (31) | 14 (43.7) | NA | (52) |
| 12 | 2007 | Tunisia | 395 | 18 - 88 | 60 | 4 (1) | 10 (2.5) | Hemodialysis | (43) |
| 13 | 2008 | Tunisia | 38 | 1 - 56 | 100 | 20 (52.6) | 17 (44.7) | Hemophilia | (42) |
| 14 | 2013 | Tunisia | 33 | 34 - 56 | 67 | 0 | 4 (12) | Hemodialysis | (50) |
Abbreviations: IDU, intravenous drug user; NA, not available.
| Year of Registry in GenBank | Country | Subtype 1a, n | Subtype 1b, n | Patient Group | Title in GenBank | |
|---|---|---|---|---|---|---|
| 1 | 2008 | Azerbaijan | 0 | 25 | IDU | Hepatitis C recombinant form 1 - 2k/1b prevalent in IDU networks in Azerbaijan |
| 2 | 2010 | Egypt | 1 | 1 | NA | HCV intrafamilial transmission in Greater Cairo, Egypt |
| 3 | 2012 | Iran | 108 | 24 | Inherited bleeding disorder | Molecular epidemiology of hepatitis C virus among patients with inherited bleeding disorders in Iran |
| 4 | 2012 | Iran | 23 | 3 | Blood donor | Genotype distribution of hepatitis C virus among Iranian blood donors, 2006 |
| 5 | 2013 | Iran | 22 | 1 | Blood donor | Genotype distribution of hepatitis C virus among Iranian blood donors, 2006 - 2008 |
| 6 | 2010 | Pakistan | 2 | 0 | NA | Hepatitis C virus subtype 1a isolate Pk-NS5B 1a non-structural protein 5B gene |
| 7 | 2011 | Pakistan | 5 | 3 | NA | NS5B genome based HCV genotyping and evolutionary analysis |
| 8 | 2005 | Tunisia | 12 | 0 | Hemophilia | Genetic variability of genotype 1 HCV strains obtained from Tunisian haemophiliacs and assessed by phylogenetic analyses in the NS5b region database |
3.3. Phylogenetic Analysis of HCV Subtype 1a
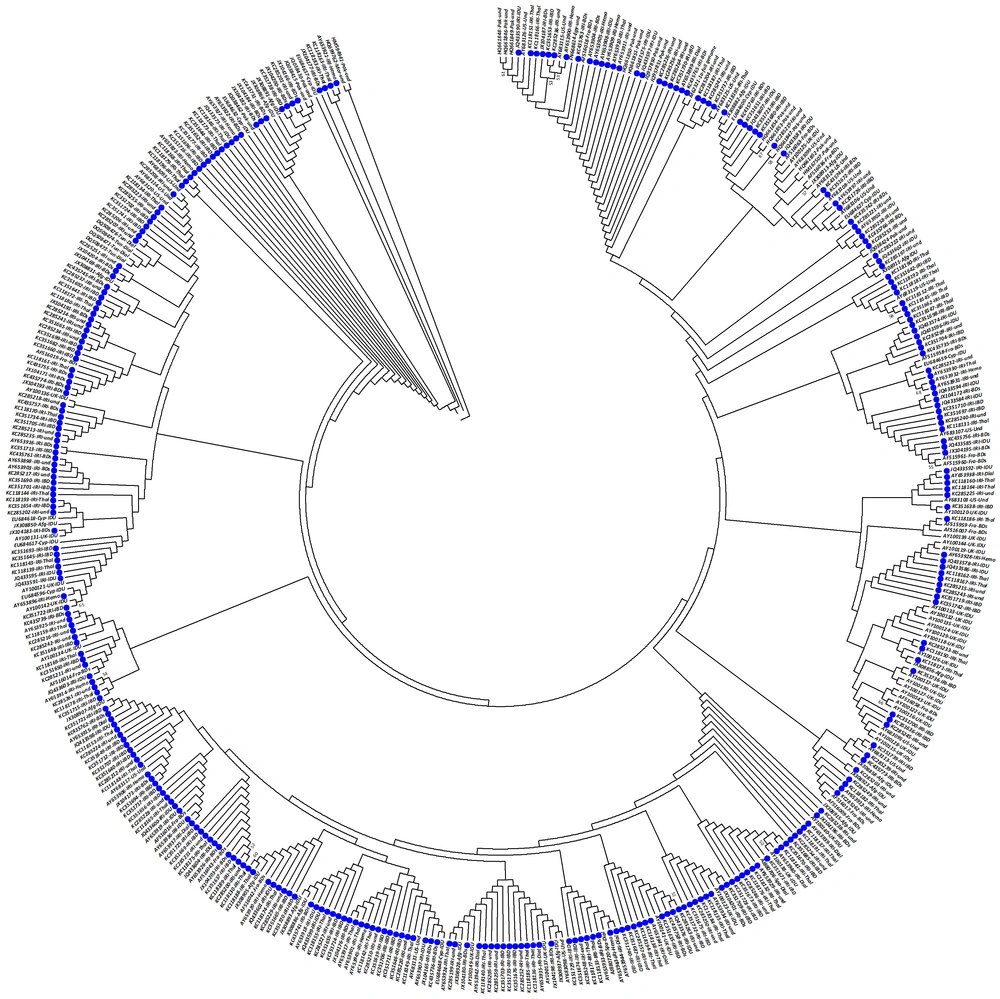
Out of 442 extracted sequences for HCV-1a, 325 (73.5%) sequences were obtained from Iranian studies. These sequences were derived from different groups including blood donors, inherited bleeding disorders, thalassemia, intravenous drug users, patients on hemodialysis, and patients without known risk factors. The phylogenetic analysis of HCV-1a demonstrated various clusters (Figure 2).
Most of the Afghan isolates clustered among Iranian isolates within the same clades. There were sequences from the UK intravenous drug users clustered with some French blood donors and some sequences from Iranian patients in the phylogenetic tree. Also, the specific sequences from Iranian patients clustering with the UK and Cypriots intravenous drug users were observed (Figure 2).
Phylogenetic Analysis of NS5B Sequences of HCV Subtype 1a, The analysis involved 442 nucleotide sequences, and codon positions included 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. Phylogenetic clusters were defined by bootstrap analysis (cut-off 50%). Values for these clusters are indicated next to the branches (values > 50% are shown). The accession number, the country of origin, and patient group are listed for all isolates. Solid blue circles indicate sequences attributed to the Iranian strains. Abbreviations of country names are as follows: Afgh: Afghanistan; Azer: Azerbaijan; Cyp: Cyprus; Egy: Egypt; Fra: France; IRI: Iran; Ita: Italy; Mor: Morocco; Ned: Netherlands; Pak: Pakistan; Spa: Spain; Tun: Tunisia; UK: the United Kingdom; USA: the United States of America. Abbreviations of patient groups are as following: BDs: blood donors; Dial: hemodialysis; IDU: intravenous drug users; IBD: inherited bleeding disorders; Hemo: hemophilia; Thal: thalassemia; LT: liver transplants; Und: undetermined.The optimal tree with the sum of branch length of 1.380 is shown, and there were a total of 187 positions in the final dataset.
It can be concluded from the tree that a proportion of the Iranian isolates clustered along with each other.
3.4. Phylogenetic Analysis of HCV Subtype 1b
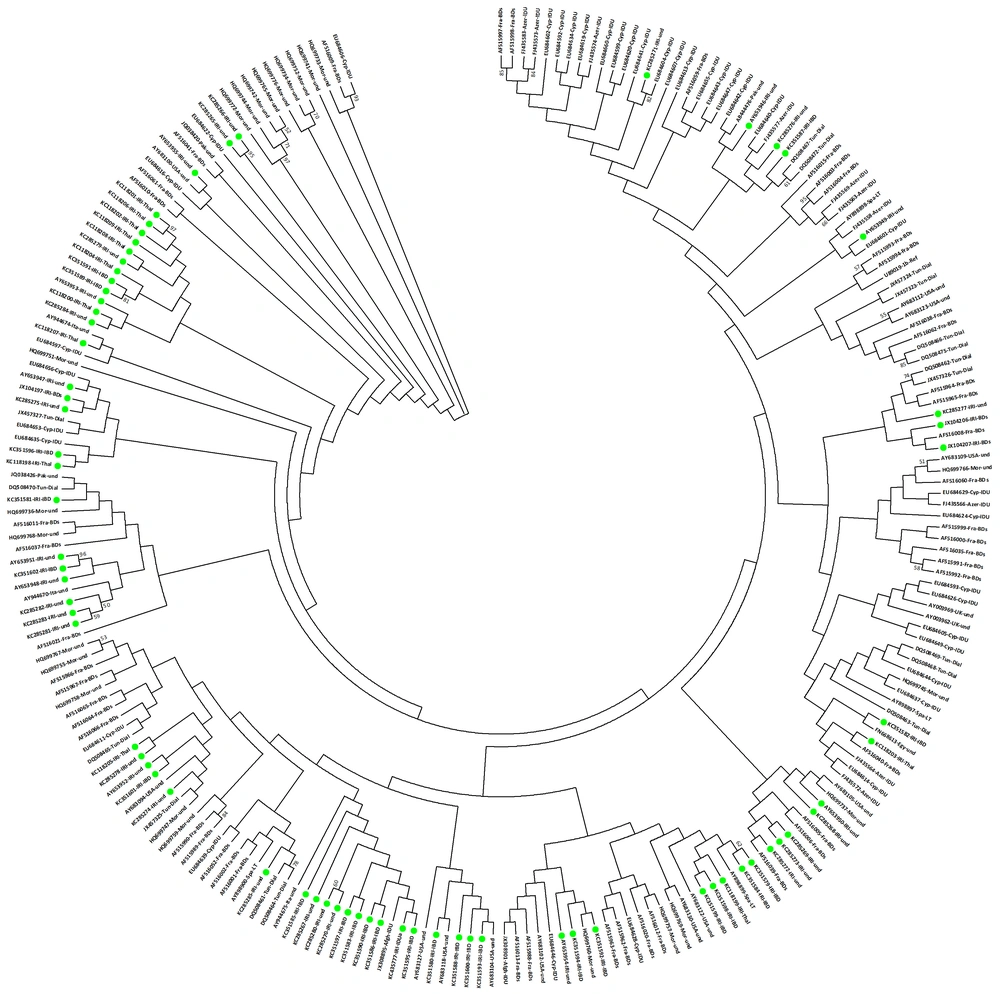
A comparison of sequences of HCV-1b is shown in the phylogenetic tree in Figure 3. Seventy one (30.6%) sequences of total 232 sequences were isolates from Iranian patients. These sequences were isolated from blood donors, inherited bleeding disorders, thalassemia, and patients without known risk factors. Based on the phylogenetic analysis, the Iranian sequences of HCV-1b had heterogeneous dispersion (Figure 3). Some of the Iranian sequences clustered with each other and some clustered with European sequences particularly sequences from France, Spain and Italy. Most likely, the HCV-1b isolates from Iranian patients may have similarities with the European ones. Also, in the phylogenetic tree there are isolates from different geographical regions which clustered together. It is likely that the subtype 1b has different origins.
Phylogenetic Analysis of NS5B Sequences of HCV Subtype 1b, The analysis involved 232 nucleotide sequences. The codon positions included 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. Phylogenetic clusters were defined by bootstrap analysis (cut-off 50%). Values for these clusters are indicated next to the branches (values > 50% are shown). The accession number, the country of origin, and patient groups are listed for all isolates. Solid green circles indicate sequences attributed to the Iranian strains. Abbreviations of country names are: Afgh: Afghanistan; Azer: Azerbaijan; Cyp: Cyprus; Egy: Egypt; Fra: France; IRI: Iran; Ita: Italy; Mor: Morocco; Ned: Netherlands; Pak: Pakistan; Spa: Spain; Tun: Tunisia; UK: the United Kingdom; USA: the United States of America. Abbreviations of patient groups are written as: BDs: blood donors; Dial: hemodialysis; IDU: intravenous drug users; IBD: inherited bleeding disorders; Hemo: hemophilia; Thal: thalassemia; LT: liver transplants; Und: undetermined. The optimal tree with the sum of branch length of 3.468 is shown, and there were a total of 203 positions in the final dataset.
4. Conclusions
Currently, HCV is considered as one of the most important viruses threatening human life. HCV has 7 major genotypes and more than 67 different subtypes. The distribution pattern of HCV genotypes is various among HCV-infected individuals which depends on different status of public health behavior and social risk factors. It is clear to all that the predominance of risk factors for HCV transmission has changed over time, from blood transfusion to intravenous drug use (24). The distribution of HCV genotypes and subtypes in Iran and other Middle Eastern and EMRO countries has a very diverse pattern. Altogether, the HCV-1 (-1a and -1b) is the predominant genotype in Iran, so that more than half of the HCV-infected patients in Iran are infected with this genotype (with a rate of 54%) (12, 18).
The genetic diversity of HCV is due to the unique characteristic of the RNA molecule. The genetic variation stems from the error-prone NS5B polymerase. As a result, different populations of viruses called “quasispecies” are produced, almost with a single mutation in each cycle of replication. The production of highly different viruses through the dynamic replication process of HCV will occur with a count of 10 trillion viruses per day (53).
The phylogenetic pattern gives valuable information about hierarchal relationships and genetic evolution. The results of the detailed phylogenetic analysis by using NS5B sequences of HCV-1a indicated that a proportion of the Iranian HCV-1a isolates was in common clades. Therefore, it can be concluded that “a proportion of Iranian HCV-1a isolates most probably has domestic origin”. In this study, most of Afghan strains have fallen into Iranian strains. These results showed that the HCV-1a sequences from Afghan patients were likely similar to the isolates from Iran. This indicates that the HCV-1a sequences from both countries are closely related to each other genetically. This may be because of the fact that the Afghanistan land was a part of Iranian territory years ago and now after occurring fled wars in Afghanistan since 1978, Iran’s border gates were opened on Afghan refugees and it provided the conditions for the large numbers of Afghans to immigrate to Iran. Perhaps this could be one of the main explanations for the HCV genotypes similarity in Afghanistan and Iran.
In Pakistan, HCV-1a was the third predominant genotype (with a rate of 4.82%). It is plausible that the most of Pakistani patients who were infected with HCV-1a, acquired this infection due to unsafe medical practices during surgeries (54). In a previous phylogenetic analysis of HCV-1a in Pakistan, the virus was identified with polyphyletic origin, and the sequences were found to be closely related to European strains (55).
Discussion surrounding HCV-1b is more sophisticated, as it has different geographical distribution patterns. HCV-1b is the third dominant genotype in Iran. Cyprus, Morocco, Tunisia, and Turkey are the countries wherein HCV-1b is predominant. The EMRO and Middle Eastern countries take diverse patterns of HCV genotype distribution, dominantly HCV-G4 in Arab countries to HCV-G3 and -G1 (-1a and -1b) in non-Arab countries (18).
As mentioned earlier, some of the HCV-1b sequences of Iranian isolates were similar to counterpart sequences from European isolates including those from France and Spain. It seems that this similarity is more likely among Iranian patients with inherited congenital bleeding disorder such as hemophilia. The prevalence of HCV among Iranian hemophilia patients is high, as it is said that the overall prevalence in these Iranian patients group is 40.8%, with a range from 13.3% to 80.5% (8, 56). It is noteworthy that in the 1980s, with the arrival of blood and blood products from France to Iran, a large number of patients, particularly hemophiliac patients, were infected with HIV and HCV. The different pattern of HCV infection in Iranian hemophiliac patients has been clearly defined. Previously, it was shown that HCV-1b is more frequently observed in Iranian hemophiliac patients than other Iranian HCV infected groups (57). This may indicate the possible similarity of HCV-1b of Iranian and European isolates and suggests an infection through blood products such as clotting factors imported to Iran.
Phylogenetic analysis of HCV genotypes and subtypes is a useful molecular method which helps scientists in every geographic region provide a substantial contribution to monitoring the virus for any purpose including HCV molecular tracing, ancestral studies, performing different genetic assessments, and guidance for any treatment decision. There are some limitations in this study: 1) the short (< 300 bp) nucleotide sequences which were used in the phylogenetic analyses, 2) HCV NS5B sequences were available from limited number of EMRO and middle eastern countries, and 3) we could not establish a proper analysis for assessment of the relationship between HCV risk factors and the phylogeny of HCV NS5B sequences. However, more studies should be conducted in next future to find more genetic relationships between these different sequences from different regions and patient groups including genetic distances for measuring genetic divergence, phylodynamic inference, and evolutionary methods to define circulating strains and molecular clock analysis for understanding the ancestral relationships.
In conclusion, the NS5B sequences of different infected-patients were phylogenetically- evolutionarily analyzed for molecular tracing of HCV-1 in Iran. The phylogenetic trees of HCV-1a and -1b according to 500 pseudo-replicates indicated many clades and codon positions with ancestral relationships of all data. Phylogenetic reconstruction of all sequences of HCV-1 pinpoints phylogenetic dispersion of most of HCV-1b of Iranian isolates among other European ones with a considerable diversity; whereas, most of Iranian HCV-1a isolates are genetically defined probably with domestic origin.