1. Background
Hepatitis C virus (HCV) infection affects between 60% and 80% of individuals, and over the course of 20 years, there is a 15% to 30% probability of developing hepatocellular carcinoma (HCC) and cirrhosis (1). Understanding the processes that underlie the infection's resolution or persistence may depend on identifying the changes in cytokine production caused by HCV. However, several studies in this field have shown a preference for host factors such as illness duration, immunological and genetic background, liver disease stage involving hepatic fibrosis and inflammation, and HCV features such as genotype and virulence (2, 3). A large number of cells, including stromal, endothelial, fibroblast, and immune cells (mast cells, T or B lymphocytes, and macrophages), secrete glycoproteins, also called cytokines, which are humoral immunomodulatory proteins (4). The intricate network of cytokines that function during the initial infection enables the synchronized and effective progression of both innate and adaptive immune responses (4). However, the HCV interacts with cytokines at various stages and eludes the immune response by triggering the cytotoxic T/T-helper (Th)-2 (5). Cytokines play a crucial role in regulating inflammation during viral infections, such as HCV infection. They also act as intercellular mediators involved in controlling the virus and causing liver damage in HCV infection (5). Multiple cellular types produce the versatile cytokine interleukin 10 (IL-10). Macrophages, primarily innate immune cells, possess a higher cytokine production capacity than adaptive immunity cells. This is because macrophages can rapidly respond to pathogens and initiate a broad range of cellular reactions. This phenomenon can also be observed in tumors injected with myeloid-derived suppressor cells (6). T-helper-2 cells produce IL-10, inhibiting other cytokines that induce inflammation (7). The IL-10 gene consists of five exons on chromosome 1 (8). Three single nucleotide polymorphisms (SNPs) were identified within the promoter area of the IL-10 gene: -1082 G/A, -819 C/T, and -592 C/A. These SNPs are consistent with the transcriptional activity of the IL-10 promoter (8) and are responsible for the variation in IL-10 production among individuals. Afzal et al. found that different IL-10 gene variations are important in determining both how quickly an infection spreads and how likely a person is to get HCV (9).
2. Objectives
Exploring genetic variations is critical to understanding the effects of genetic background in HCV infection. The main objective of this study was to explore the links between a variation in the IL-10 gene promoter region -1082 (rs1800896) and HCV infection in a sample of Iraqi patients. Furthermore, we investigated possible correlations between the different genotypes of the SNP and the clinic pathological characteristics of the patients.
3. Methods
3.1. Study Groups
This study obtained ethical approval from the Iraqi Ministry of Health, Medical City Department, Training, and Human Development Center (No. 31630). Blood samples were collected from 180 participants who were referred to the Digestive and Liver Diseases Teaching Hospital in Baghdad Medical City. The study comprised 100 individuals with HCV infection and 80 healthy controls. All participants provided their consent by signing an informed consent form.
3.2. Inclusion and Exclusion Criteria
Inclusion criteria for the patient group were a positive result for the HCV antibody test and/or the HCV Nucleic Acid Test (NAT). Inclusion criteria for the control group were a negative result for both the HCV antibody and HCV NAT tests. Participants with a prior medical record or diagnostic findings indicating the presence of autoimmune disorders, or factors contributing to cirrhosis such as alcohol consumption, obesity, and diabetes, were excluded from both the patient and control groups.
3.3. Molecular Study
The study involved molecular profiling of the IL-10 gene promoter region -1082 (G/A; rs1800896) in all enrolled subjects. Five milliliters of blood samples were collected from the peripheral vein by puncturing a sterile vein. Specimens were preserved at -20°C until the IL-10 rs1800896 A/G polymorphism was detected using molecular biology techniques. DNA was extracted from whole blood using the ReliaPrepTM blood g-DNA mini-prep systems. Specific primers (Table 1) were designed using GeneRunner and used for the amplification of the IL-10 promoter region encompassing the rs1800896 SNP by polymerase chain reaction (PCR). The PCR products were analyzed by agarose gel electrophoresis (1.5%) and imaged with a gel documentation system. The PCR products were sent to Macrogen Company (Korea) for sequencing, which uses the ABI3730XL automated DNA sequencer for Sanger sequencing. The sequence reads were aligned with the reference DNA sequence obtained from the National Center for Biotechnology Information (NCBI), and the genotypes were determined.
| SNP | Primer | Primer Sequences (5’ to 3’) | Annealing Temperature (°C) | Product Size (bp) |
|---|---|---|---|---|
| IL-10 rs1800896 | IL-10 forward | TGTAAAACGACGGCCAGTCAGGGAGGATGAGTGATTTG | 60 | 853 |
| IL-10 reverse | CAGGAAACAGCTATGACGTGTTCCAGGCTCCTTTAC |
Abbreviations: SNP, single nucleotide polymorphism; IL-10, interleukin 10.
3.4. Statistical Analysis
SPSS-26, a specialized statistical software program, was utilized to conduct an analysis of the data in terms of both its demographic and genetic characteristics. Normality was checked using the Shapiro-Wilk test. Quantitative variables were compared between groups using the independent t-test or Mann-Whitney U test. Qualitative variables were analyzed using the Kruskal-Wallis test. The allele and genotype differences between the two groups, association with the disease, and the genetic model action were assessed by calculating odds ratio (OR) and confidence interval (CI) values using WinPepi software. The data were presented as the average value with or without the standard deviation. Statistics were considered significant when the P-value was less than or equal to 0.05.
4. Results
4.1. Patients’ Characteristics
The laboratory and demographic characteristics of HCV patients and healthy controls are outlined in Table 2. The variables include number, age, gender distribution, and method of acquiring the virus. Among the patients, there were 46 (46%) men and 54 (54%) women, compared with 28 (35.0%) men and 52 (65.0%) women in the control group. The causes of disease varied, including unknown (38), surgery (16), dentist (19), cancer (7), thalassemia (7), blood transfusion (7), and dialysis (6). The mean age of the patients was 38.8 ± 13.3 years, while the mean age of the control group was 34.5 ± 14.5 years.
| Demographic Characteristic | Patients (N = 100) | Controls (N = 80) | P-Value |
|---|---|---|---|
| Gender | > 0.001 b | ||
| Male | 46 (46) | 28 (35.0) | |
| Female | 54 (54) | 52 (65.0) | |
| Cause of infection | - | ||
| Not infected | 0 (0) | 80 (100) | |
| Unknown | 38 (38) | 0 (0) | |
| Surgery | 16 (16) | 0 (0) | |
| Dents | 19 (19) | 0 (0) | |
| Cancer | 7 (7) | 0 (0) | |
| Thalassemia | 7 (7) | 0 (0) | |
| Blood transfusion | 7 (7) | 0 (0) | |
| Dialysis | 6 (6) | 0 (0) | |
| Age (y) | 38.8 ± 13.3 | 34.5 ± 14.5 | 0.019 c |
Abbreviations: P-value, probability value; SD, standard deviation.
a Values are expressed as No. (%) or mean ± SD.
b Was considered statistically highly significant.
c Variables were considered statistically very significant.
4.2. Genotyping of the Patients and Controls
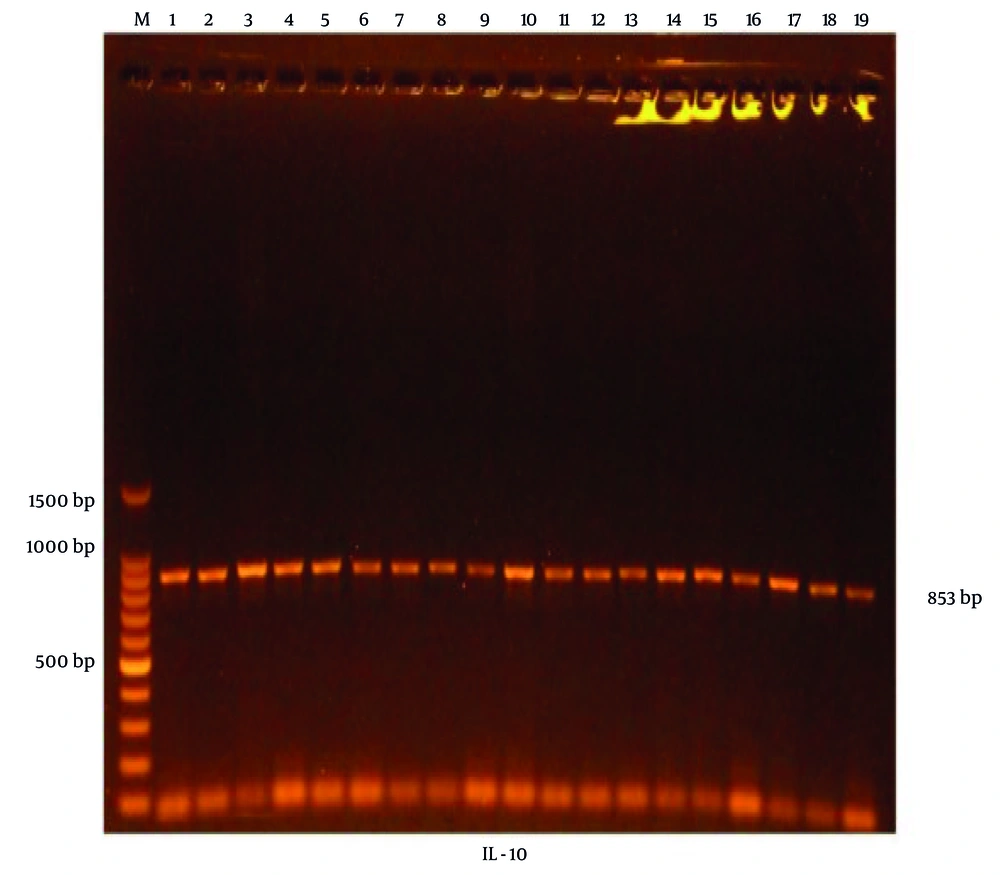
After genomic DNA extraction, the samples were used as DNA templates in PCR to amplify the specific area containing rs1800896 of IL-10. The PCR products were then electrophoresed on a 1.5% agarose gel, stained with Ethidium Bromide, and imaged using a gel documentation system. As indicated in Figure 1, all PCR products displayed a specific electrophoretic band representing a 583 base pair fragment length.
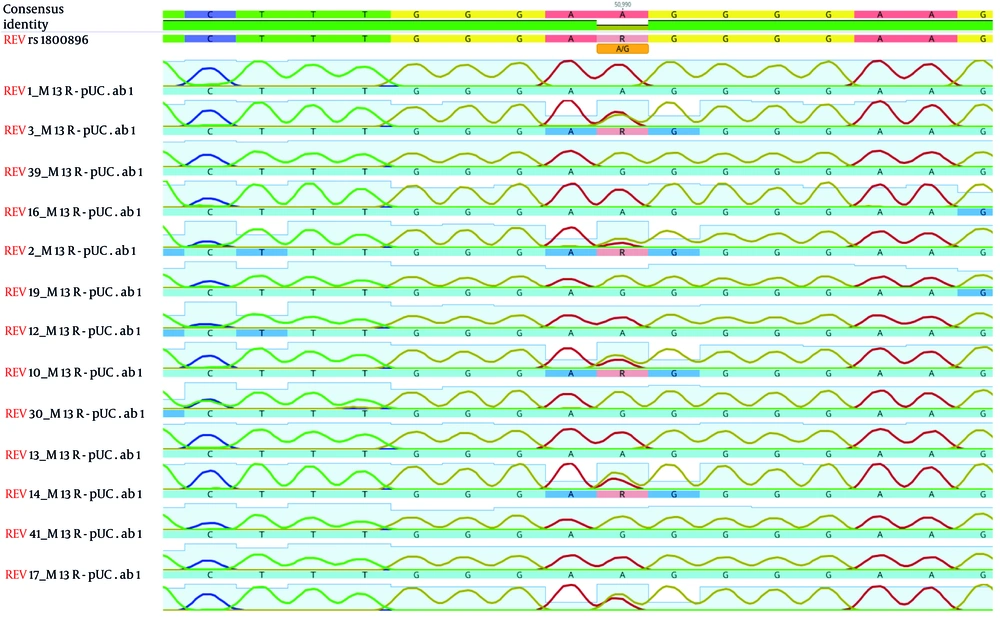
Genotypes of the samples were determined by sequencing in Macrogen (Corporation, Korea) as shown in Figure 2.
Performing Sanger sequencing to determine the genotype of the rs1800896 single nucleotide polymorphism (SNP) in the interleukin 10 (IL-10) gene. A solitary "A" peak indicates the presence of an A homozygous allele. A solitary "G" peak suggests the presence of a G homozygous allele. The existence of the "A" and "G" peaks suggests the presence of the A/G heterozygous allele.
The genotypes of patients and controls were analyzed. The wild-type AA frequency was 33% in patients compared to 48.7% in controls (OR = 0.52, CI = 0.28 - 0.94, P-value = 0.046). The heterozygous AG frequency was 50% in patients compared to 30% in controls (OR = 2.33, CI = 1.26 - 4.31, P-value = 0.009). The difference between the two groups for the AG genotype was very significant, with a high-risk OR. However, the homozygous GG genotype had a frequency of 17% in both groups, displaying no difference (P-value = 0.56, OR = 0.76, CI = 0.36 - 1.60). For allele frequencies, a non-significant association was observed as the differences were insignificant (P = 0.28) (Table 3).
Abbreviations: SNP, single nucleotide polymorphism ; HCV, hepatitis C virus; CI, confidence interval; OR, odds ratio; NS, non-significant.
a Values are expressed as No. (%).
b P < 0.05 was considered statistically significant.
c P < 0.01 was considered statistically very significant.
The link between the IL-10 rs1800896 polymorphism and disease-associated risk was revealed by calculating three models: Dominant, recessive, and over-dominant. The AG + GG dominant model and the AG over-dominant model were shown to be significantly different. These connections are considered risk factors with OR of 1.93 (95% CI; 1.60 - 3.52, P = 0.046) and 2.33 (95% CI; 1.26 - 3.31, P = 0.009), respectively. The results showed no significant changes in the recessive model (OR = 0.76, 95% CI; 0.36 - 1.60, P = 0.56), as shown in Table 4.
Abbreviations: CI, confidence interval; OR, odds ratio; NS, non-significant; Ref, reference; N, number; P, probability.
a P < 0.05 was considered statistically significant.
b P < 0.01 was considered statistically very significant.
Table 5 displays the demographic characteristics of both the study and control groups. The factors include genotype, count, age, gender distribution, and cause of virus acquisition. No statistically significant correlation exists between genotypes, age, and gender in cases and controls, as indicated by P-values of 0.1, 0.07, and 0.06, respectively. The genotypes among HCV patients were distributed as follows: AA (33, 33%), AG (50, 50%), and GG (17, 17%). The genotypes in the control group were distributed as follows: AA (39, 48.7%), AG (24, 30%), and GG (17, 21.3%).
| Demographics Characteristic | rs1800896 | P-Value | |||||
|---|---|---|---|---|---|---|---|
| Controls | Patients | ||||||
| AA | AG | GG | AA | AG | GG | ||
| Age (y) | 35.4 ± 14.6 | 33.6 ± 13.3 | 33.7 ± 14.8 | 40.0 ± 16.7 | 37.2 ± 17.7 | 41.1 ± 18.0 | 0.07, NS |
| Gender | 0.06, NS | ||||||
| Male | 10 (35.7) | 13 (46.4) | 5 (17.8) | 15 (32.6) | 24 (52.2) | 7 (15.2) | |
| Female | 29 (55.8) | 11 (21.1) | 12 (23.1) | 18 (33.3) | 26 (48.2) | 10 (18.5) | |
| Cause of infection | 0.02 b | ||||||
| No. | 39 (48.7) | 24 (30.0) | 17 (21.3) | ||||
| Unknown | - | - | - | 10 (26.3) | 21 (55.3) | 7 (18.4) | |
| Surgery | - | - | - | 6 (37.5) | 7 (43.8) | 3 (18.7) | |
| Dental | - | - | - | 8 (42.1) | 9 (47.4) | 2 (10.5) | |
| Cancer | - | - | - | 2 (28.6) | 4 (57.1) | 1 (14.3) | |
| Thalassemia | - | - | - | 4 (57.1) | 2 (28.6) | 1 (14.3) | |
| Blood transfusion | - | - | - | 2 (28.6) | 3 (42.8) | 2 (28.6) | |
| Dialysis | - | - | - | 1 (16.7) | 4 (66.6) | 1 (16.6) | |
Abbreviations: P, probability; NS, non-significant.
a Values are expressed as mean ± SD or No. (%).
b Variables were considered statistically significant.
5. Discussion
The aim of our study is to investigate the IL-10 cytokine gene polymorphism rs1800896 and its possible links to various aspects of viral infection and vulnerability to chronic HCV infection. The results showed that genotype can influence the incidence and susceptibility to the disease. The findings indicated that individuals with the AG/GG genotype were more susceptible to HCV infection than those with the AA genotype. Approximately 60 - 80% of individuals with HCV infections are unable to clear the virus on their own, resulting in the development of chronic, recurrent infections. Since the virus does not directly damage the liver, the pathophysiology of a long-term infection with HCV seems to be associated with the immune system locally. One of the most important immunological responses that plays a role in the pathophysiology of HCV infection is the generation of cytokines, which can stimulate or restrict growth and have anti- or pro-inflammatory effects (10). Numerous investigations have demonstrated that this immunological reaction and the course of the HCV virus are significantly influenced by host genetic factors (11). The age group between 20 and 70 years is the most common. There are statistically significant differences in demographic parameters between HCV patients and healthy controls (Table 1). It was reported that infection with HCV at older ages might increase the risk of developing chronic HCV infection (12). In our study, the mean age was different between the patient and control groups, but the difference was not statistically highly significant (P = 0.019) and might be a source of slight bias. Furthermore, a recent study discovered that although women are less likely to develop the disease than men, women experience symptoms more frequently than men (13). This is due to the fact that, as Pathak (2022) notes, estrogen, a hormone present in women, interacts with liver cells to shield them from the HCV (14). Yet, this resistance is diminished when women's hormone levels fall, but these reports are not in agreement with other reports (15, 16). In the current study of the IL-10 gene polymorphism rs1800896, which is situated in the gene's promoter region, the frequency of the wild-type AA genotype in the control group was higher than that in the patient group, highlighting its evident protective role. There was also a statistically significant difference in the frequencies of the heterozygous AG genotype, as its frequency in the patient group is higher than in the control group, indicating a risk factor for the disease. Nonetheless, allele frequencies were not significantly different between the patient and control groups. It is worth noting that IL-10 can be produced by many lymphocytes and myeloid cells of different types, and it is possible to stimulate more than one group of cells producing IL-10 during a single infection (17, 18).
The main histocompatibility antigens produced by cells are downregulated by IL-10, which diminishes the immunological response to an antigen (19). It is possible that SNPs of the IL-10 gene within the promoter region affect IL-10 secretion and its peripheral anti-inflammatory effects, contributing to combating disease progression as well as attenuating its transcription rate (20, 21). This might predispose individuals with polymorphisms in the IL-10 gene to chronic HCV infection. The results in Table 4 show three genetic models: Dominant, recessive, and over-dominant. According to the dominant model, compared to the homozygous AA genotype, having at least one variant allele (G) in the genotypes (AG or GG) raises the risk of HCV (22). The study in this publication demonstrates that individuals with the AG or GG genotype have a 1.93-fold increased risk of HCV compared to those with the AA genotype. According to the recessive model, contracting HCV is likely linked to having two copies of the mutant allele (GG). However, results did not find a significant association between the GG genotype and HCV compared to the AG or AA genotypes, which is in agreement with the report by Zschocke et al. (23). According to Gemmell and Slate, in the heterozygote advantage, also known as the over-dominant model, the heterozygote genotype is more suited than either of the homozygous genotypes (24). Consistent with this model, our findings showed that the probability of HCV infection in people with the AG genotype is 2.33 times higher than in people with the AA or GG genotypes. Thus, in the studied population, the AG genotype is more susceptible to HCV than the AA or GG genotypes. These results and findings by others suggest that the IL-10 rs1800896 polymorphism may influence an individual's susceptibility to HCV through different genetic models (5, 25). Table 5 also shows a comparison between the three genotypes AG, AA, and GG in each group of patients and the control group with demographic characteristics in terms of gender and age. There was no statistically significant difference between the control and patient groups regarding the genotypes to link the risk of HCV infection to the age or gender parameters. These observations were not consistent with a study conducted in China, where a statistically significant result was found between HCV and age (12). The outcomes of the current study align with other studies reporting a notable correlation between HCV infection and the IL-10 rs1800896 gene polymorphism (26-28). On the other hand, some studies have denied the correlation between IL-10 rs1800896 gene polymorphism and HCV infection (29). Additionally, research conducted in Egypt involving 220 healthy individuals and 440 patients with HCV infection found no association between HCV infection and the IL-10 rs1800896 gene polymorphism (30). Finally, in the present study, a limited number of patients referred to the Digestive and Liver Diseases Teaching Hospital in Baghdad Medical City was used as a sample population for the study. Furthermore, only one of the IL-10 polymorphisms was considered here. These were limitations of the study, reflecting the need for further research using a larger sample size and more polymorphisms.
5.1. Conclusions
A higher possibility of developing an HCV infection was detected in individuals with the AG genotype, as per the current study. In contrast, those with the AA genotype exhibit a protective effect in healthy individuals. The results indicate that the IL-10 rs1800896 polymorphism may have a notable impact on HCV infection, thus warranting further investigation in genetics and medicine.