1. Background
Hepatitis B virus (HBV) infection remains a major public health problem in the world. Although effective vaccine is available, there are still about a million new infections annually and about 240 million chronic infections worldwide (1). The clinical outcome of HBV infection varies from spontaneous recovery to persistent infection that may increasingly progress to liver cirrhosis (LC) and/or hepatocelluar carcinoma (HCC) (2). Studies have shown that the polymorphism of the genes correlated with the host immunological function plays an important role in the progression of chronic HBV infection (3). Human leukocyte antigen (HLA) class II gene polymorphism has been confirmed to be associated with the outcome of HBV infection by some researchers (4-6). Class II transactivator (CIITA) is the master regulator of HLA-II gene expression. It regulates whether HLA-II gene expression or the expression level of HLA-II molecule so as to affect the outcome of HBV infection (7).
HBV exhibits remarkable host specificity and liver tropism. Until now, the mechanism of HBV particles entry into the host hepatocytes is still enigmatic (8), although more than a dozen host-binding proteins, such as preS1, preS2, and S domains, have been identified in the last two decades. In 2012, Yan et al. (9) demonstrated that sodium taurocholate cotransporting polypeptide (NTCP) is a functional receptor for HBV, and it is crucial for binding to the receptor-binding region of the pre-S1 domain of the L protein of HBV and critically contributed to NTCP-mediated HBV infection.
So far, there are a few reports about the association between single nucleotide polymorphisms (SNPs) of CIITA NTCP and chronic HBV infection. Therefore, we conducted a case-control study to investigate the influence of CIITA and NTCP gene variants on chronic HBV infection as well as the disease progression.
2. Objectives
This study aimed to determine the association of CIITA and NTCP gene variants with HBV infection as well as the disease progression.
3. Methods
3.1. Study Subjects
A total of 671 unrelated Han Chinese were recruited for this study between October 2012 and April 2014 (Table 1). Written informed consent was obtained from all the subjects, and the study was performed with the approval of the ethics committee of the first affiliated hospital of Fujian Medical University. Chronic HBV infection subjects were divided into three groups: a) Chronic hepatitis B group (CHB); b) LC group; and c) HCC group. The diagnosis of CHB was based on seropositivity for HBsAg and persistently elevated levels of serum aminotransferase for longer than six months, in accordance with the criteria issued by the Chinese society of hepatology and the Chinese society of infectious diseases in 2010 (10). LC and HCC patients were diagnosed according to the criteria as previously described (11). Clearance subjects were negative for HBsAg and positive for both HBsAb and HBcAb, while they were normal in the indicators of liver function. The subjects who were positive for HBsAb and negative for HBcAb were excluded because of their possible history of vaccination. The healthy subjects had normal indicators of liver function and negative HBV serological markers. Subjects who had any other type of liver diseases such as autoimmune liver disease and alcoholic liver disease and those infected with hepatitis C virus and human immunodeficiency virus were excluded from the study.
| Characteristics | Healthy Subjects (n = 170) | Clearance Subjects (n = 199) | Chronic HBV Infection Subjects (n = 305) | ||
|---|---|---|---|---|---|
| CHB (n = 169) | LC (n = 68) | HCC (n = 68) | |||
| Gender | |||||
| Malea | 77 (45.3) | 94 (47.2) | 105 (62.1) | 53 (77.9) | 58 (85.3) |
| Femalea | 93 (54.7) | 105 (52.8) | 64 (37.9) | 15 (22.1) | 10 (14.7) |
| Ageb | 45.5 ± 10.1 | 45.4 ± 11.3 | 34.1 ± 9.7 | 52.1 ± 10.9 | 56.2 ± 12.2 |
| HBsAg (-/+) | 170/0 | 199/0 | 0/169 | 0/68 | 0/68 |
| HBsAb (-/+) | 170/0 | 0/199 | 169/0 | 68/0 | 68/0 |
| HBeAg (-/+) | 170/0 | 199/0 | 59/110 | 50/18 | 49/19 |
| HBeAb (-/+) | 170/0 | 199/0 | 110/59 | 18/50 | 19/49 |
| HBcAb (-/+) | 170/0 | 0/199 | 0/169 | 0/68 | 0/68 |
Abbreviations: CHB, Chronic Hepatitis B; HCC, Heptocellular Carcinoma; LC, Liver Cirrhosis.
aValues are expressed as No. (%).
bValues are expresse as Mean ± SD.
3.2. SNPs Selection
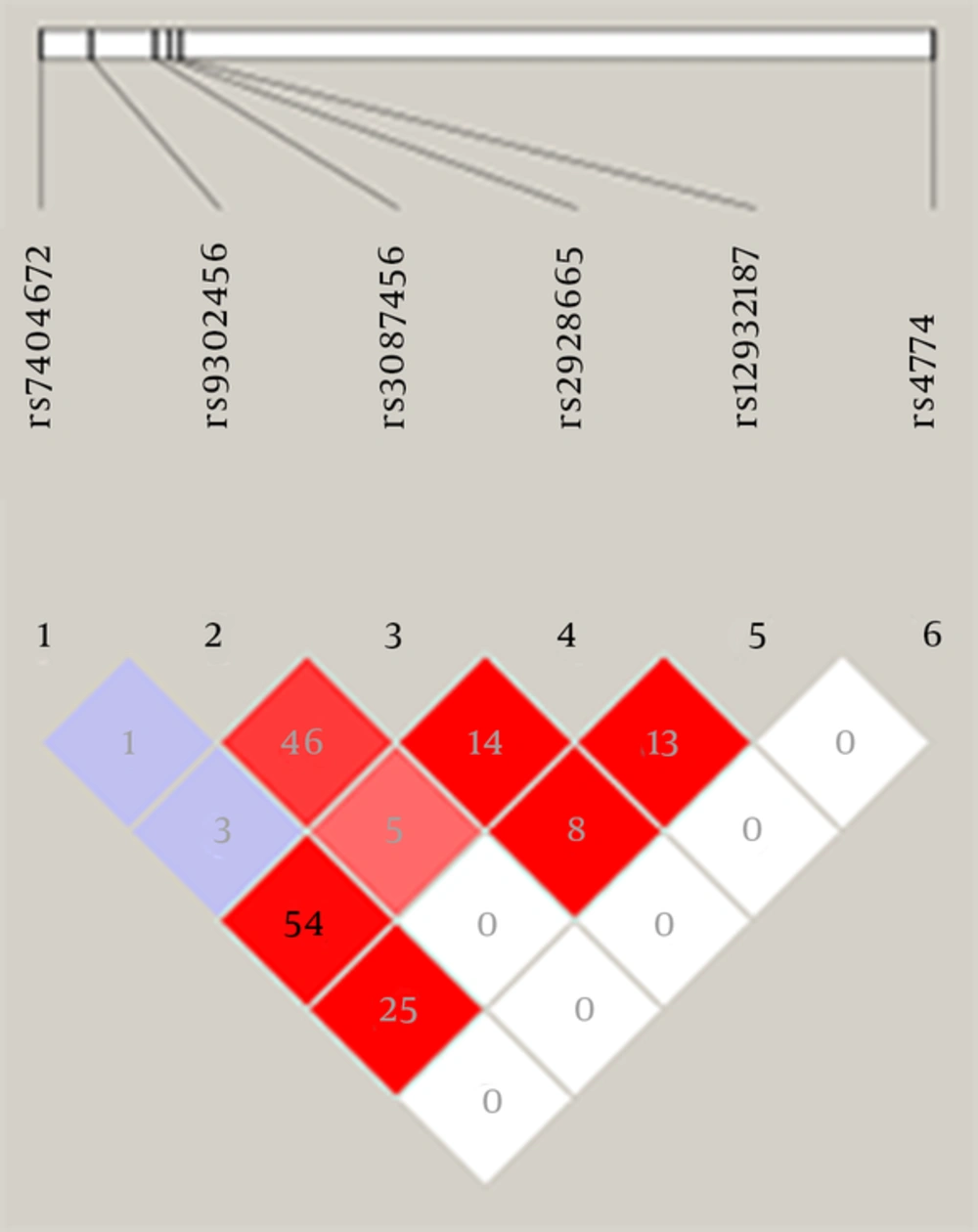
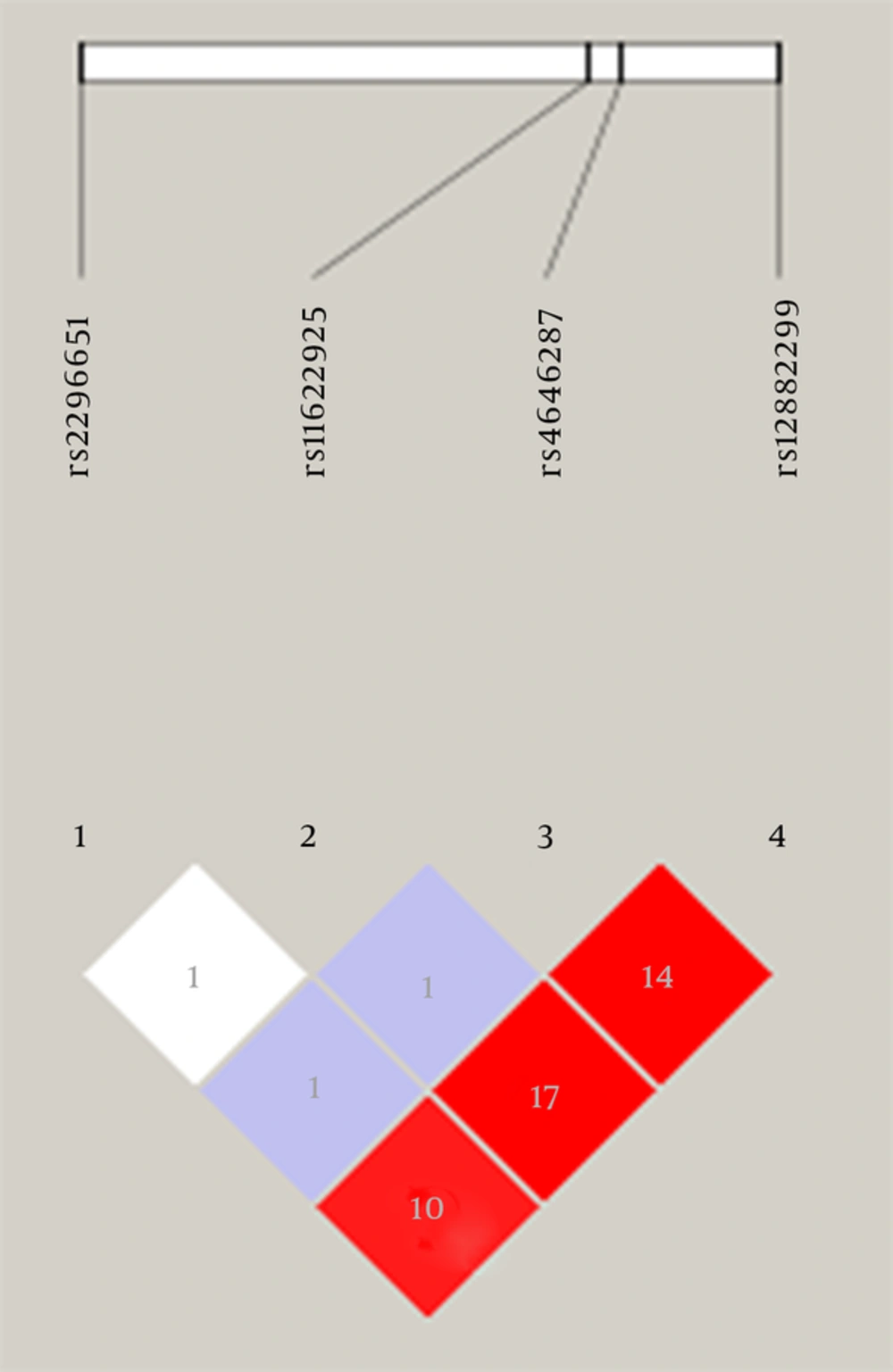
We chose six SNPs for CIITA, five of which (rs7404672, rs9302456, rs3087456, rs12928665, and rs12932187) located in the promoter, and the sixth one (rs4774) in exon 11. For NTCP, we selected tagger SNPs using the genotype data of the sample of Han Chinese in Beijing, China, from the International HapMap Project database using Haploview 4.2 and the criteria: minor allele frequency (MAF) ≥ 0.02; and R2 ≥ 0.8, which was according to HapMap Data Rel 28 PhaseII + III, August 10, on NCBI B36 assembly, dbSNP b126. Four SNPs (rs2296651, rs11622925, rs4646287, and rs12882299) were identified near the region of NTCP gene on chromosome 14.
3.3. SNPs Genotyping
Genomic DNA was extracted from 2 ml of EDTA-anticoagulated peripheral blood samples using a TIANamp DNA Kit (Tiangen Biotech Co., Ltd., China) according to the manufacturer’s instructions. The SNPs genotyping work was performed using an improved multiplex ligation detection reaction (iMLDR) technique (12), which was recently developed by Genesky Biotechnologies Inc. (Shanghai, China). The iMLDR is an improved multiplex SNP discrimination technology based on the traditional ligation detection reaction (LDR). Ten SNPs were amplified in a system with multiplex PCR. The amplified products had been purified with ExoI/SAP before they were applied to the template for a subsequent ligase detection reaction. In a connection reaction, each site included two 5’ terminal allele-specific probes, followed by a 3’ terminal fluorescently labeled specific probe. All primers, probes, and labeling oligos were designed by and ordered from Genesky Biotechnologies Inc. The primer and probe information in two mixtures is described in Tables 2 and 3, respectively.
| Primer Name | Primer Concentration | Primer Sequence |
|---|---|---|
| rs7404672F | 1 μM | TTTCCCTGCATTCCTACCAGCTA |
| rs7404672R | 1 μM | GCTTTTTGGTGAAAGAGCACTGG |
| rs9302456F | 1 μM | AGGTGCCAAAGTGCATCCTCTG |
| rs9302456R | 1 μM | GTGACTGCAGCTGCCTGGTACA |
| rs3087456F | 1 μM | TGTTGAAGGTTCCCCCAACAGA |
| rs3087456R | 1 μM | CCCAGCTCAGAAGCACACAGC |
| rs12928665F | 2 μM | CAGGGGTCTGGACAAGGAGGTT |
| rs12928665R | 2 μM | CGTGGAAGGCAACTGTGCTTTTA |
| rs12932187F | 1 μM | AGGGTGTGCCCCTGAAGAAGTC |
| rs12932187R | 1 μM | TTAAGGCTGCACCCAACCACAC |
| rs4774F | 1 μM | TATGGCCTGCAGGATCTGCTCT |
| rs4774R | 1 μM | GCCTTGCTCAGGCTCTGGAC |
| rs2296651F | 1 μM | GTCCCTGCTAGAAACTTGCTTGTTG |
| rs2296651R | 1 μM | TGGTAGCAGCACTGGGACAAAG |
| rs11622925F | 1 μM | CAGGGGTCTGGACAAGGAGGTT |
| rs11622925R | 1 μM | CGTGGAAGGCAACTGTGCTTTTA |
| rs4646287F | 1 μM | TCTCCCCAGTTTGGAAGGATGA |
| rs4646287R | 1 μM | AGAGTTTCCCAGCACCCACTCC |
| rs12882299F | 2 μM | CATTGCCACATGGCACATTCAC |
| rs12882299R | 2 μM | GCCATGCAAATTGGGCAGATTA |
| Probe Name | Probe Concentration | Probe Sequence |
|---|---|---|
| rs7404672FC | 1 μM | TTCCGCGTTCGGACTGATATCCTACCA |
| GCTAAGCCCCCTTTACTAC | ||
| rs7404672FT | 1 μM | TACGGTTATTCGGGCTCCTGTCCTACC |
| AGCTAAGCCCCCTTTACCAT | ||
| rs7404672FP | 2 μM | AATCCATCAGCAGGTCCAGGTTTTTTT |
| rs9302456FC | 1 μM | TCTCTCGGGTCAATTCGTCCTTGGATG |
| TCACTTGCTCTGCTCAGAGTCC | ||
| rs9302456FT | 1 μM | TGTTCGTGGGCCGGATTAGTGGATGTC |
| ACTTGCTCTGCTCAGAGTCT | ||
| rs9302456FP | 2 μM | TGCACYGACACAGGCATGGTTTTTTT |
| rs3087456RA | 1 μM | TACGGTTATTCGGGCTCCTGTCCACAC |
| TCCCTTAAGCCCTCACT | ||
| rs3087456RG | 1 μM | TTCCGCGTTCGGACTGATATCCACACT |
| CCCTTAAGCCCTCACC | ||
| rs3087456RP | 2 μM | ACACCTCTGAAATTAATTTCACTTCCT |
| TACTGTTTT | ||
| rs12928665RA | 1 μM | TACGGTTATTCGGGCTCCTGTGCCCGA |
| AAGTCTGACTTCTAGAACGTT | ||
| rs12928665RG | 1 μM | TTCCGCGTTCGGACTGATATGCCCGAA |
| AGTCTGACTTCTAGAACATC | ||
| rs12928665RP | 2 μM | TCGGTGCTGATACATGGTTCATACTTTT |
| rs12932187FC | 1 μM | TGTTCGTGGGCCGGATTAGTGCCCCTGA |
| AGAAGTCGTTTACATTGTC | ||
| rs12932187FG | 1 μM | TCTCTCGGGTCAATTCGTCCTTGCCCCT |
| GAAGAAGTCGTTTACATTGTG | ||
| rs12932187FP | 2 μM | GAGTCAATTTTCCTGGAGTGTACAATGTT |
| rs4774RC | 1 μM | TGTTCGTGGGCCGGATTAGTCGCTTCCAG |
| CTCCTCGATGC | ||
| rs4774RG | 1 μM | TCTCTCGGGTCAATTCGTCCTTCGCTTCC |
| AGCTCCTCGATGG | ||
| rs4774RP | 2 μM | CGTCTAGGATGAGCAGAACGCGTTTTT |
| rs2296651RA | 1 μM | TGTTCGTGGGCCGGATTAGTGGATGCCAAAAT |
| GTCCAACTCTGGTT | ||
| rs2296651RG | 1 μM | TCTCTCGGGTCAATTCGTCCTTGGATGCCAAA |
| ATGTCCAACTCTGATC | ||
| rs2296651RP | 2 μM | CACCATCCTCAATGTGGCCTTTTT |
| rs11622925FC | 1 μM | TTCCGCGTTCGGACTGATATGTGGCCACTGGC |
| GATGGTGC | ||
| rs11622925FT | 1 μM | TACGGTTATTCGGGCTCCTGTGTGGCCACTGG |
| CGATGGTGT | ||
| rs11622925FP | 2 μM | TGGGACATCTGGCCACTCACTTTTTTT |
| rs4646287RC | 1 μM | TTCCGCGTTCGGACTGATATAAGCAGAAATCA |
| GCAAGGGCACG | ||
| rs4646287RT | 1 μM | TACGGTTATTCGGGCTCCTGTAAGCAGAAATC |
| AGCAAGGGCACA | ||
| rs4646287RP | 2 μM | CTCCTGGAGACRCAGCACACTTTTTTTT |
| rs12882299RC | 1 μM | TCTCTCGGGTCAATTCGTCCTTTCTATTTTTA |
| TTGCTTTGTTGTCCAAGGTTG | ||
| rs12882299RT | 1 μM | TGTTCGTGGGCCGGATTAGTTCTATTTTTATT |
| GCTTTGTTGTCCAAGGCTA | ||
| rs12882299RP | 2 μM | TGATTGGTATAATTTTATTTTTGTTTTTTGCA |
3.4. Statistical Analysis
The Hard-Weinberg equilibrium of genotypes was evaluated by using Arlequin 3.5. Linkage disequilibrium (LD) was assessed by Haploview 4.2 using frequencies obtained from the healthy subjects. Odds ratios (OR) and 95% confidence intervals (CI) were calculated on the basis of the binary logistic regression analysis (adjustment for gender and age). All the statistical analyses were performed by SPSS software version 20.0 and P < 0.05 in a two sided test was considered statistically significant.
4. Results
4.1. H-W Equilibrium Test and LD Analyses
The genotype frequencies of CIITA and NTCP genes in the control subjects were confirmed to be in H - W equilibrium (P > 0.05), except for rs7404672, rs3087456, and rs11622925 in the clearance subjects and rs2296651 in the healthy subjects. LD analyses performed on all individuals from the healthy group showed that the R2 value of each pair of SNPs was less than 0.80 (Figures 1 and 2).
4.2. CIITA and NTCP Loci Polymorphisms and Chronic HBV Infection
The dominant model was selected for further study because some SNPs had lower frequencies of homozygous genotype mutation. CIITA rs9302456 CT+TT genotype was significantly higher in the chronic HBV infection than healthy subjects (OR = 2.24, 95%CI: 1.17 - 4.29). NTCP rs2296651 AG + AA genotype decreased significantly in the chronic HBV infection than healthy subjects (OR = 0.41, 95%CI: 0.23 - 0.74), whereas rs12882299 CT+CC genotype increased significantly in the chronic HBV infection compared to the healthy subjects (OR = 1.97, 95%CI: 1.27 - 3.07). By using clearance subjects as control group, we found that rs12882299 CT + CC genotype increased the risk of HBV infection (OR = 1.71, 95%CI: 1.13 - 2.58) (Table 4).
| SNP | Healthy Subjects | Clearance Subjects | Chronic HBV Infection Subjects | Pa OR (95%CI) | Pb OR (95%CI) |
|---|---|---|---|---|---|
| rs7404672 | |||||
| CC | 98 | 121 | 171 | ||
| CT | 56 | 61 | 119 | 0.620 | 0.405 |
| TT | 16 | 17 | 15 | 1.11 (0.74 - 1.64) | 1.17 (0.80 - 1.71) |
| rs9302456 | |||||
| CC | 155 | 171 | 260 | ||
| CT | 13 | 27 | 44 | 0.015 | 0.356 |
| TT | 2 | 1 | 1 | 2.24 (1.17 - 4.29) | 1.28 (0.75 - 2.19) |
| rs3087456 | |||||
| GG | 144 | 164 | 250 | ||
| AG | 25 | 27 | 54 | 0.282 | 0.471 |
| AA | 1 | 8 | 1 | 1.34 (0.78 - 2.28) | 1.19(0.73 - 1.94) |
| rs12928665 | |||||
| GG | 71 | 80 | 114 | ||
| AG | 69 | 87 | 144 | 0.250 | 0.469 |
| AA | 30 | 32 | 47 | 1.26 (0.84 - 1.88) | 1.51(0.78 - 1.68) |
| rs12932187 | |||||
| CC | 62 | 72 | 107 | ||
| GC | 74 | 93 | 148 | 0.997 | 0.843 |
| GG | 34 | 34 | 50 | 0.99 (0.66 - 1.50) | 0.96 (0.65 - 1.41) |
| rs4774 | |||||
| GG | 126 | 152 | 229 | ||
| GC | 41 | 46 | 68 | 0.910 | 0.876 |
| CC | 3 | 1 | 8 | 0.97 (0.62 - 1.52) | 1.03 (0.67 - 1.59) |
| rs2296651 | |||||
| GG | 139 | 171 | 278 | 0.003 | 0.168 |
| AG | 19 | 28 | 27 | 0.41 (0.23 - 0.74) | 0.67 (0.37 - 1.19) |
| AA | 12 | 0 | 0 | ||
| rs11622925 | |||||
| CC | 130 | 155 | 222 | 0.361 | 0.252 |
| CT | 38 | 37 | 76 | 1.23 (0.79 - 1.94) | 1.29 (0.84 - 1.99) |
| TT | 2 | 7 | 7 | ||
| rs4646287 | |||||
| CC | 138 | 165 | 251 | 0.826 | 0.929 |
| CT | 29 | 30 | 51 | 0.95(0.58 - 1.57) | 1.02(0.63 - 1.67) |
| TT | 3 | 4 | 3 | ||
| rs12882299 | |||||
| TT | 56 | 65 | 71 | 0.003 | 0.011 |
| CT | 75 | 98 | 174 | 1.97 (1.27 - 3.07) | 1.71 (1.13 - 2.58) |
| CC | 39 | 36 | 60 |
aHealthy subjects versus chronic HBV infection subjects.
bClearance subjects versus chronic HBV infection subjects.
4.3. CIITA and NTCP Loci Polymorphisms with HBV Progression
We were interested in the possible association between the polymorphisms of CIITA and NTCP genes and the progression of chronic hepatitis B. Therefore, we further analyzed the differences in 10 SNPs genotype distributions by using CHB as control group. As can be seen, among HCC group, LC group, and CHB group, the distribution of CIITA and NTCP genotype frequencies showed no significant difference in the dominant genetic model (Table 5).
| SNP | CHB | LC | HCC | Pa OR (95%CI) | Pb OR (95%CI) |
|---|---|---|---|---|---|
| rs7404672 | |||||
| CC | 89 | 43 | 39 | ||
| CT | 73 | 21 | 25 | 0.520 | 0.752 |
| TT | 7 | 4 | 4 | 0.78 (0.36 - 1.66) | 1.14 (0.50 - 2.58) |
| rs9302456 | |||||
| CC | 143 | 56 | 61 | ||
| CT | 26 | 11 | 7 | 0.091 | 0.515 |
| TT | 0 | 1 | 0 | 2.52 (0.86 - 7.37) | 1.57 (0.40 - 6.21) |
| rs3087456 | |||||
| GG | 140 | 54 | 56 | ||
| AG | 29 | 13 | 12 | 0.160 | 0.412 |
| AA | 0 | 1 | 0 | 2.04 (0.75 - 5.56) | 1.63 (0.50 - 5.27) |
| rs12928665 | |||||
| GG | 61 | 27 | 26 | ||
| AG | 85 | 28 | 31 | 0.572 | 0.876 |
| AA | 23 | 13 | 11 | 0.80 (0.37 - 1.72) | 1.06 (0.46 - 2.46) |
| rs12932187 | |||||
| CC | 57 | 23 | 27 | ||
| GC | 87 | 33 | 28 | 0.670 | 0.338 |
| GG | 25 | 12 | 13 | 0.83 (0.37 - 1.88) | 0.65 (0.27 - 1.55) |
| rs4774 | |||||
| GG | 126 | 51 | 52 | ||
| GC | 38 | 14 | 16 | 0.997 | 0.758 |
| CC | 5 | 3 | 0 | 1.00 (0.43 - 2.31) | 1.15 (0.46 - 2.86) |
| rs2296651 | |||||
| GG | 151 | 65 | 62 | 0.507 | 0.717 |
| AG | 18 | 3 | 6 | 0.54 (0.09 - 3.33) | 1.33 (0.28 - 6.21) |
| AA | 0 | 0 | 0 | ||
| rs11622925 | |||||
| CC | 123 | 50 | 49 | 0.881 | 0.069 |
| CT | 42 | 16 | 18 | 1.07 (0.43 - 2.65) | 2.41 (0.93 - 6.22) |
| TT | 4 | 2 | 1 | ||
| rs4646287 | |||||
| CC | 141 | 55 | 55 | 0.669 | 0.517 |
| CT | 27 | 12 | 12 | 1.23 (0.47 - 3.23) | 0.71 (0.24 - 2.03) |
| TT | 1 | 1 | 1 | ||
| rs12882299 | |||||
| TT | 35 | 19 | 17 | 0.137 | 0.498 |
| CT | 102 | 35 | 37 | 0.52 (0.22 - 1.23) | 0.71 (0.27 - 1.90) |
| CC | 32 | 14 | 14 |
aCHB versus LC.
bCHB versus HCC.
5. Discussion
Recently, genome-wide association studies in Japanese and Korean populations have shown that gene variants at HLA-DP and HLA-DQ are associated with chronic HBV infection (4, 5, 13), and the result has been verified in many studies (14-16). CIITA plays a pivotal role in the immune response by controlling HLA-II gene expression, and it is considered to be an important candidate gene in immune diseases (17). In 2007, Zhang et al. (18) indicated that CIITA gene promoter IV upstream -1350C/T (rs12928665) and -944G/C (rs12932187) were associated with chronic HBV infection. It is likely that due to differences in geographic distribution of study populations, our study was not consistent with the previous study. Swanberg et al. (19) reported that CIITA gene promoter III upstream -168A/G (rs3087456) was associated with increased susceptibility to rheumatoid arthritis and multiple sclerosis. The explanation was that A→G substitution of promoter III of CIITA gene was associated with lower induction of HLA-II gene. The rs4774 (Gly500Ala) located in exon 11 has been confirmed by many studies to be associated with rheumatoid arthritis and multiple sclerosis (20, 21). However, our study did not identify the association of rs3087456 and rs4774 with chronic HBV infection.
Human NTCP protein includes 349 amino acids and contains a putative seven- or nine-transmembrane domain with a predicted topology of N-termianl extracellular and C-terminal intracellular ends (22). Several SNPs that alter the transporter activity of NTCP have been reported (23, 24). Yan et al. (25) found, in subsequent studies, that a homozygotic mutation of rs2296651 (p.Ser267Phe) on the fourth exon 4 of the NTCP gene can significantly reduce its activity. A mutation in p.Ser267Phe reduced bile acid absorption and also affected its combination with the HBV Pre-S1 protein, leading to the reduced efficiency of HBV infection. The efficiency of HBV infection and transport of taurocholic acid increased by 70% in HepG2 cells transinfected with the wild genotype and p.Ser267Phe mutant genotype at 1:1 compared to HepG2 cells transinfected with the mutant genotype. It is reasonable to speculate that heterozygous individuals may also be susceptible to HBV infection. Because our research only identified the rs2296651 AA genotype in healthy subjects, and there was a significant difference only between healthy subjects and the chronic HBV infection group, our results also confirmed the speculation. It should be noted that the p.Ser267Phe variant is located beyond the amino acids 157 to 165 of NTCP, which have been demonstrated to be critical for HBV entry into the hepatocyte (26). Possibly the p.Ser267Phe mutation changed the topology of NTCP and impaired HBV entry into the hepatocyte.
Peng et al. (27) and Su et al. (28) had published NTCP gene polymorphisms about chronic HBV infection when our manuscript was preparing. Peng et al. (27) found that A allele of rs2296651 decreased the risk of chronic HBV infection, which was consistent with our findings. We found that the frequency of the A allele of rs2296651 in the healthy subjects of this study (12.6%) is higher than the previously reported frequency in Koreans (3.1%) (24) and Chinese-Americans (7.5%) (23). The frequency is also higher than that previously documented in Chinese (4.7%) (25). These may reflect the differences of the ethnic and geographic distributions of this variant. Su et al. (28) found that rs7154439 AA genotype increased viral clearance because rs7154439 and rs12882299 both are located in the 5’ flanking region of NTCP gene and showed a weak LD (R2 = 0.255). Our results show that rs12882299 increased the risk of chronic HBV infection. We further found that rs7154439 and rs11622925 showed a strong LD (R2 = 0.884). However, our study did not identify rs11622925 associated with chronic HBV infection.
In conclusion, our study indicated that rs2296651 was negatively correlated with chronic HBV infection, and rs12882299 and rs9302456 could increase the risk of chronic HBV infection. However, considering the poor repeatability of the existing studies, a large sample size, particularly in multicenter research, is needed to verify the correlation between CIITA and NTCP gene variants and chronic HBV infection.