1. Background
The Hepatitis B virus (HBV) is one of the main infectious agents that causes liver disease and is one of the most critical health problems worldwide (1). This medical issue may lead to an acute or chronic infection, and around 350 to 400 million individuals around the world are chronically infected with HBV (2). Over 90% of new-borns, and up to half of all younger children between age 1 and 5 with infection, will develop a chronic infection (3, 4). Although some infected adults with HBV could recover, 5% to 7% of individuals develop a chronic HBV infection (5, 6).
The prevalence of chronic HBV infection is classified to three levels: low (< 2%), intermediate (2% to 7%) and high (> 8%) endemicity, and according to the world health organization (WHO) and the centers for disease control and prevention (CDC), Iran’s HBV infection prevalence is intermediate (7, 8).
Evidence from previous studies proposed that host genetic factors, including a variety of immune cytokines and receptor genes, especially single nucleotide polymorphisms play a major role in determining immunization, clinical course, and recovery from HBV infection (9-11).
The CC chemokine receptor 5 (CCR5) is a gene that encodes the CCR5 protein and is situated on the short arm of chromosome 3. One of the mutant types of the gene produces a non-functional receptor that is called CCR5Δ32, which results from deletion of a specific sequence of 32 base-pairs (12, 13).
The CCR5 plays a role in T cell proliferation and activation by binding to its ligand RANTES (CCL5) and additionally works as the principal co-receptor for macrophage (M)-tropic strains of Human Immunodeficiency Virus type 1 (HIV-1) (14, 15). The latest studies have demonstrated the protective role of homozygous for the CCR5 Δ32 genotype against HIV infection, yet in relation to heterozygous CCR5 ∆32 (∆32/wt), the results have been practically controversial (16, 17).
This homozygous genotype is known to positively effect recovery from HBV infection and provides genetic epidemiological evidence for the role of CCR5 in the immune response to HBV (18). The CCR5 Δ32 genotype frequency varies depending on the geographic area as its mean frequency is around 9.1% in the European population, and is much lower in Asia (19, 20).
2. Objectives
In this study, the researchers aimed at assessing the protective effect of CCR5 Δ32 genotype in subjects, who have recovered from HBV infection, as well as subjects that developed chronic HBV infection, in Birjand, Iran.
3. Methods
3.1. Study Participants
This case-control study consisted of 60 patients with chronic HBV infection (patient group) and 120 patients, who have previously recovered from HBV infection, spontaneously (control group).
Samples were collected from hepatitis comprehensive plan in Birjand, the capital of the South Khorasan province of Iran, during years 2013 and 2014. The comprehensive plan consisted of 5235 individuals aged 15 to 70 years. This plan included 786 (15%) subjects with positive hepatitis B core antibody (anti-HBc) as subjects with recovered HBV infection and 81 (1.6%) subjects with hepatitis B surface antigen (HBsAg) seropositivity as chronic HBV infected subjects (21).
Subjects with chronic hepatitis B (patient group) were defined as individuals having HBsAg positive test results for at least 6 months before the study entrance. The HBsAg negative/ HBc antibody positive individuals with repeatedly normal serum alanine aminotransferase (ALT) levels were considered as controls, and had recovered from HBV infection spontaneously.
The HBV infected patients with HDV or HCV co-infection, autoimmune hepatitis, malignity, alcohol history, and toxic agent use were excluded from the study.
This study was conducted after obtaining approval from the ethics committee of Birjand University of Medical Sciences, Iran (approval code: Ir.bums.REC.1395.58).
3.2. DNA Extraction
For each sample, 10 mL of whole blood was collected in tubes containing EDTA anticoagulant. The tubes were centrifuged at 2500 rpm for 15 minutes to separate the buffy coat layer and stored at -20°C. The DNA was extracted by salting out method and then DNA concentration and Purity were analyzed by Nanodrop Spectrophotometer (Bio Tek, USA EPOCH) and DNA integrity was determined by visual inspection for degradation by electrophoresis (there was no long smear and excess band).
3.3. PCR and Gel Electrophoresis
DNA was analyzed for the presence or absence of the CCR5 Δ32 deletion by using polymerase chain reaction (PCR) amplification. The primer set CCR5F, 5-CAAAAAGAAGGTCTTCATTACACC-3 (situated on the third exon, nucleotides 519 to 542) and CCR5R, 5-CCTGTGCCTCTTCTTCTCATTTCG-3 (situated on the third exon, nucleotides 684 to 707) with Genbank accession number U54994 was used for amplification of wild and mutant types (22).
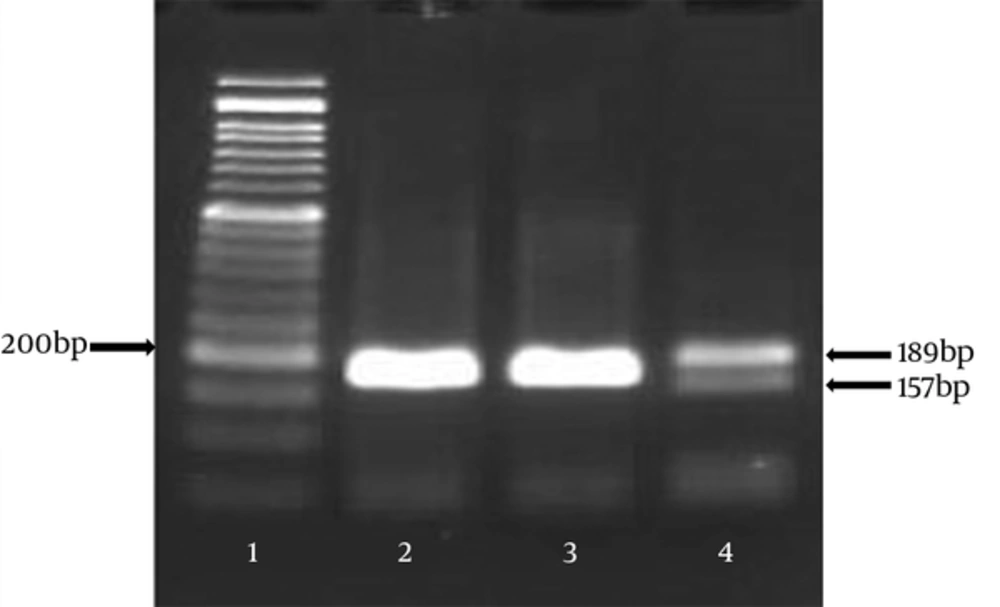
To determine the CCR5 Δ32 mutation in CCR5, primers were designed in a manner to cover the complete deleted region in the CCR5 gene. Based on the size of the selected region (relevant primer designs), PCR amplification resulted in two products: 189 bp representing the wild type allele and 157 bp representing the CCR5-Δ32 allele (Figure 1).
Samples were amplified in a final volume of 25 µL, containing final concentration of 1X standard Taq reaction buffer, 2 mM MgCl2, 0.1 M of both primers, 200 µM of each dNTP, and 0/625 Unit Taq polymerase.
Polymerase chain reaction (PCR) was performed by the Mastercycler system (Germany Eppendorf 5345) and the profile consisted of a first denaturation step of 3 minutes at 94°C, second denaturation followed by 32 cycles of 35 seconds at 94°C, 30 seconds at 58°C for annealing step, and 30 seconds at 72°C; and a final elongation step of 3 minutes at 72°C.
After PCR, amplified DNA was observed on 2% agarose gel, by gel electrophoresis in TBE buffer and a 50-bp ladder was also used to be compared with the PCR fragments (189 bp for the wild-type allele and 157 bp for the 32 bp-deletion allele) and run at 95 volt for 1 to 2 hours.
3.4. Sequencing of the PCR Product
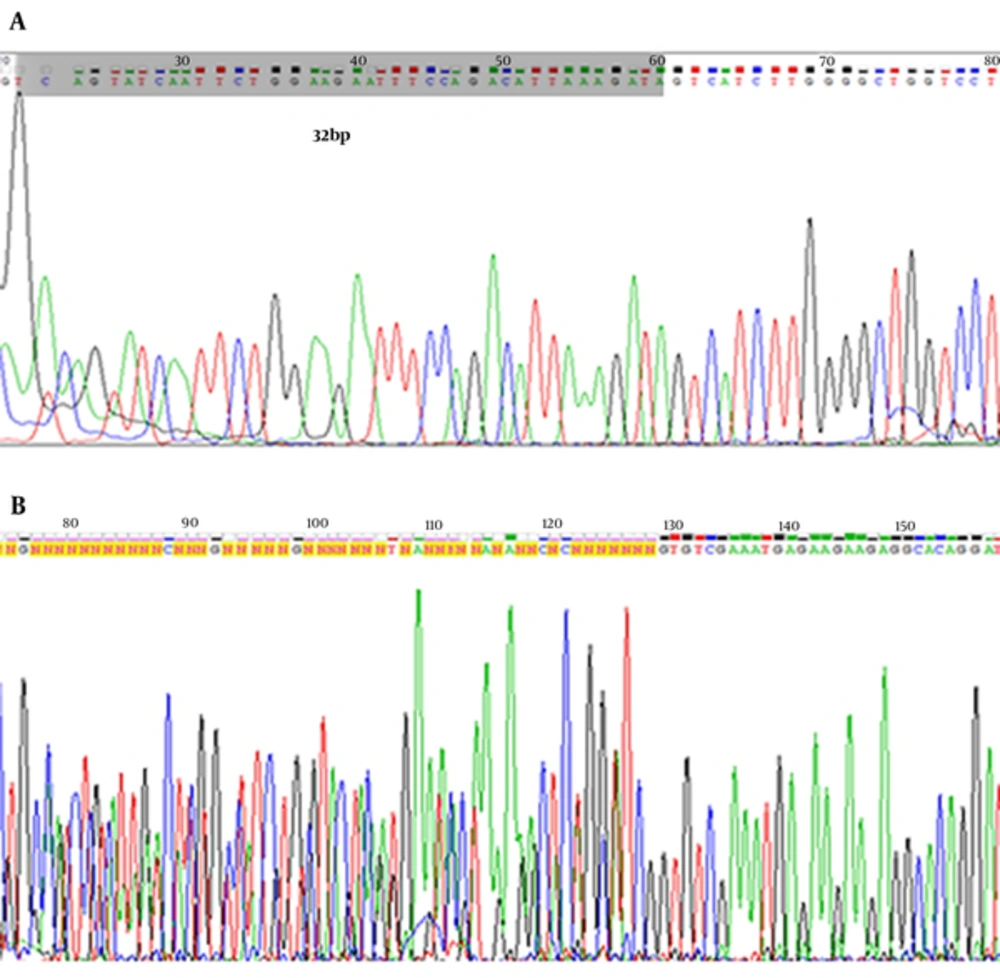
The heterozygous CCR5 Δ32 mutation sample and two wild-type homozygous were processed through direct sequencing using an applied biosystems 3730/3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA). Sequence alignment analysis was performed using the BLAST and Chromas Lite 2.1.1 Software.
3.5. Statistical Analysis
Statistical analysis was done using SPSS 17.0 software. Variables were analyzed using the Chi-square test and P values less than 0.05 were considered statistically significant.
4. Results
Characteristics of the 2 groups are compared in Table 1. There was no significant difference between the control and patient groups in age and gender.
| Variable | Controls | Patients | P Valuea |
|---|---|---|---|
| Total No. (within group) | 120 | 60 | |
| Age, y, mean ± SD | 47.5 ± 13.0 | 45.1 ± 13.7 | 0.164 |
| Gender, No. (%) | |||
| Male | 65 (54.2) | 31 (51.7) | 0.751 |
| Female | 55 (45.8) | 29 (48.3) | 0.751 |
aComparison of values between patient and control group.
The researchers found only one heterozygous CCR5 ∆32 mutation in the control group and absolutely no homozygous CCR5 ∆32 mutation in either the patient or control group (Figure 1).
This displays that there was no significant visible correlation between carrying the mutation, and the possibility of recovery from HBV infection in the city of Birjand (Table 2).
| Gene Profiles | Controls (N = 120) | Patients (N = 60) | P Valuea |
|---|---|---|---|
| CCR5 (wt/∆32) genotype, No. (%) | 1 (0.8) | - | 1.000 |
| CCR5 (wt/wt) genotype, No. (%) | 119 (99.2) | 60 (100) | 1.000 |
| CCR5 (∆32 ) allele frequency | 0.0042 | - | 1.000 |
| CCR5 (wt) allele frequency | 0.9958 | 1 | 1.000 |
Abbreviations: wt, Wild-Type; No, Number of Subjects.
aComparison of values between patient and control group
The heterozygous CCR5 ∆32 mutation sample and 2 wild-type homozygous were confirmed by the sequencing method. Electropherogram image of CCR5 alleles is shown in Figure 2.
5. Discussion
According to the current results, the CCR5 ∆32 allele frequency is very low in both those who have previously recovered from HBV and HBV persistent individuals; in Birjand’s population only one heterozygous individual was detected among the tested samples, so there was no apparent correlation between carrying the mutation and the possibility of recovery from HBV infection in the city of Birjand. This result is in agreement with the study carried out by M.D. Omrani et al. (2009) in the North-Western region of Iran. Their result showed that the frequency of the CCR5 ∆32 allele in heterozygote and homozygote was 0.0211 and zero, respectively (23).
Another finding that confirms the current results is evaluation of CCR5 ∆32 and CCR2-64I frequency in southern Iranian normal population as reported by M. Gharagozloo et al. Their results indicate that CCR5 ∆32 allele exists in southern Iranian individuals at a very low frequency (0.1221).
It’s mean frequency in other parts of Asia including Pakistan, India, Punjab, Saudi Arabia, Yemen, China, Japan, and Thailand is very low (nearly zero), whereas in the European population, it is about 9.1% and there is probably historical evidence that genetic admixtures, migrations, and plague epidemics could be responsible for this significant diversity between Asia and Europe (20).
As was proved, this mutation has a protective role against chronic HBV infection and HIV, and also leads to a non-functional receptor that could modify disease in Rheumatoid arthritis patients, so it is concluded that the population of Birjand has greater hereditary susceptibility to chronic HBV infections, HIV, and Rheumatoid arthritis, compared to Europeans in terms of the CCR5 ∆32 mutation (13, 14, 18).
However, based on linkage disequilibrium information, the mutation age is estimated between 700 to 3500 years old, and the origin of this mutation is likely to be Europe (24). Since Iran’s geographical location is between Asia and Europe, this low frequency of CCR5 ∆32 allele may be derived from admixture with the populations of Europe, even though the consequence of parallel autonomous mutations is also possible.
In conclusion, because of no significant visible correlation between carrying the mutation, and the possibility of recovery from HBV infection in the city of Birjand, it seems the protective effect of this mutation is absent in Birjand’s population. This population, similar to other parts of Iran and nearby Asian countries, has greater hereditary susceptibility to chronic HBV infection compared to European populations in terms of the CCR5 ∆32 mutation.