1. Background
Hepatitis viruses are one of most important concerns in the patients on hemodialysis (HD) who are at high risk for blood-borne infections due to prolonged vascular access and the potential for exposure to infected patients and contaminated equipment (1, 2). Screening and isolation for finding of patients with HBV and HCV infection are routine in HD units and these infections may coincide with additional causative hepatitis agents. In 1995 and 1996, different isolates of the same new enveloped positive-stranded flavivirus-like RNA viruses, with a genomic size of about 9.3 kb, were isolated by two independent research groups and named GB virus C (GBV-C) and hepatitis G virus (HGV), respectively (3-5). GBV-C/HGV is transmitted mainly by blood contamination and occasionally, sexual transmission. GBV-C has up to 95% sequence homology with HGV and therefore, the two are now considered the same virus (6).
Open reading frame (ORF) of this RNA virus encodes a polyprotein with about 2900 amino acids length. The polyprotein of GBV-C is cleaved which by proteases into structural proteins (e.g. Core, E1, and E2) and nonstructural proteins (e.g. NS2, NS3, NS4A, NS4B, NS5a, and NS5b) (4, 5). There are reports on prevalence of HGV in Iranian patients and normal population.
In 2005 and 2008 at investigation of GBV-C/HGV infection in Iranian patients with chronic renal failure, in patients with thalassemias, and in intravenous drug users (IVDUs) detected HGV infection with relatively intermediate frequency (7, 8).
In other report from Iran (9), a rate of 28.6% of GBV-C was reported among a group of patients with chronic hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, kidney transplant, patients on HD, and volunteer blood donors and 40% co-infection of GBV-C was also reported among patients with HCV.
In addition, Zahedi et al. found that half of those with positive marker of HGV infection (HGV RNA) were cirrhotic, whereas only 9.5% of those without positive marker of HGV infection were cirrhotic (10). Moreover, molecular evaluation of HGV and HCV in patients with chronic renal failure in Iran detected HGV-RNA in 17.74% of the patients and in 3% of blood donors (11).
Finally, the overall prevalence of HGV infection among HIV-positive subjects with parenteral and sexual exposure (including 52 IDUs and 30 cases with heterosexual orientation) was estimated at 10.97%, with no statistically significant difference between the two groups (13.5% among IDUs vs 6.7% among the sexual cases) (12).
While the NS3 or helicase region is not considered to be ideal for genotyping of the HGV, 5’ UTR and E2 information help to better identifying genotype and characteristics of HGV isolates (13, 14). Analyzing of NS3 region of HGV was reported in some countries such as Spain, Germany, China, the United States, and the United Kingdom (15-18).
2. Objectives
Review literature on Iranian HGV isolates showed no report on sequencing and phylogenetic analysis of NS3 sequence; information about this gene in Iranian HGVs was incomplete and mostly had referred to 5’ non-coding region (NCR) or E2 sequence. Therefore, the aims of present study were to investigate the sequence and phylogenetic features of Iranian NS3 gene of GBV-C/HGV obtained from patients on HD in Tehran and to compare them with other reported sequences from the other geographical regions of world.
3. Materials and Methods
3.1. Preparation of Nucleic Acids From Human Plasma Samples and RNA Extraction
Five-milliliter blood samples were obtained from three HGV-PCR-positive patients, who confirmed their infection by nested PCR test using primer from putative 5’UTR of our previous project, with various medical histories (data not shown) from four Tehran army hospitals with HD facilities. Samples were centrifuged immediately at the unit, and the separated plasma was stored at -20℃. Total RNA was extracted from 10-mL plasma samples using the guanidine is othiocyanate-phenol-chloroform method according to the manufacturer’s instructions.
3.2. Reverse Transcriptase-PCR
Reverse transcription was performed using random primer and Moloney murine leukemia virus (MMLV) reverse transcriptase (Fermentas Co, Germany) for synthesis of cDNA. The NS-3 fragment was amplified using nested PCR with Pfu DNA polymerase. The first primer pair, GB-C-s1 and GB-C-a1, included GB-C-S1 (5’-GACGTTGGTGAGATCCCCTT-3’) for forward and GB-C-a1 (5’-CGAAGTTTCCTGTGTACCC-3’) for reverse (3). The second primer pair included, a2 (5’-TAGAGAGCGCGTCTGTCGCAC-3’) and s2 (5’-TGGGCATGGTATCCCCCTCGAGC-3’), which was nested within the first set to give increased specificity and sensitivity. For both the first and second rounds, denaturation at 94°C for five minutes was followed by 40 cycles of amplification at 94°C for 45 seconds, 53°C for 45 seconds, and 72°C for 75 seconds. A prolonged incubation, at 72°C for seven minutes, was added at the end to ensure full extension of all products. PCR products were analyzed by 1.5% agarose gel electrophoresis and TAE buffer with 100 bp DNA Ladder in 80 V, and bands were visualized with ethidium bromide staining under UV-detector.
3.3. Sequencing and Analyzing Sequencing Data
Amplified fragments were prepared for sequencing without further purification and directed for sequencing to Takapouzist Co, Iran. In order to view and analyze quality of sequencing results, we took advantages of Finch TV software (Geospiza Inc, Seattle, USA).
3.4. Registering, Blasting and Alignment of Sequences
After registering of sequences in NCBI, data directed to nucleotide similarity BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) for evaluation of similar NS3 gene sequences reported before in different geographical region and we selected the closest hits and summarized them in informative tables. Obtained sequences were aligned by using ClustalX and analyzed in Bioedit software version 7.7.9 (mbio, Inc, North Carolina, USA). In addition, distance tree of Iranian partial NS3 genes from NCBI Blast results, based on fast minimum evolution by maximum sequence difference 0.75, was constructed.
3.5. Constructing the Phylogenetic Tree
For phylogenetic tree, nucleotide sequences of NS3 gene were retrieved from GenBank (http:// ncbi.nlm.nih.gov/). After alignment Iranian S3 gene of HGV with other sequences, redundant sequences, very short sequences and areas with ambiguous alignment or containing poly-N stretches were excluded from the analyses. MEGA 6 software packages used for construction of phylogenetic tree by using neighbor joining method (19).
4. Results
4.1. Electrophoresis and Sequencing Results
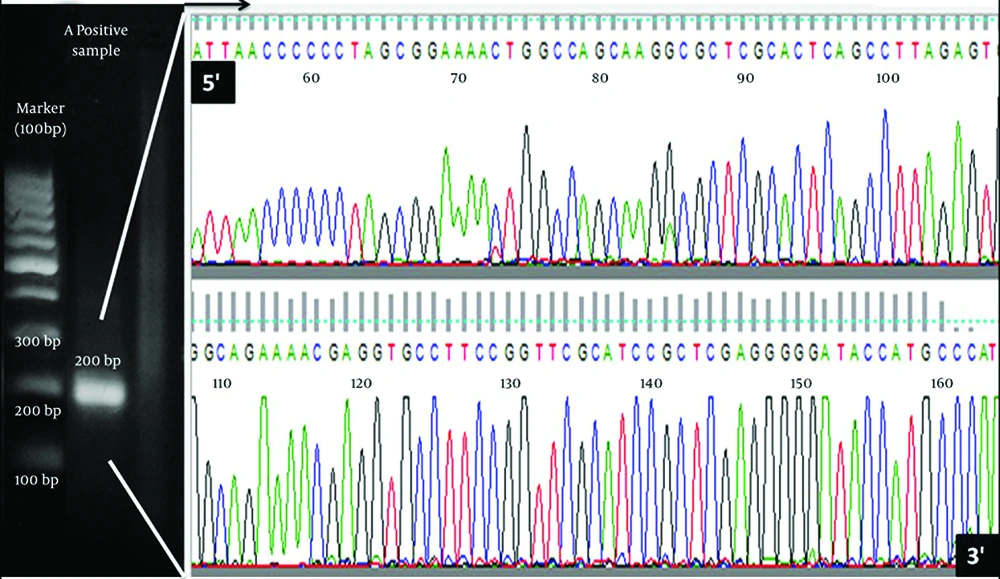
HGV NS3 fragments were amplified from total RNA extracted from three positive samples and after visualizations of 200 bp bond in gel electrophoresis, amplified partial fragments were delivered to company for direct sequencing. Sequencing NS3 gene data quality was confirmed by Finch TV software (Geospiza Inc, Seattle, USA). An electrophoresis result and chromatogram for obtained sequences as candidate of sequenced samples are demonstrated in Figure 1.
4.2. Blasting and Sequence Alignment Results
Nucleotide sequences of partial NS3 gene of Iranian GBV-C were used as entry data to NCBI BLAST search for finding probable most similar sequences. BLAST result revealed that most similar sequences to Iranian NS3sequences were those from Europeans countries (Spain, Germany, and the United Kingdom) and the United States. Most similar hits belongs to Spain (94%, maximum score 213) and Germany-USA (94%, maximumscore 207). Details of similarity are summarized in Table 1. In addition, pairwise identity calculation of Iranian sequences showed 96.4% identity between IR1 and IR2, 94.9% between IR1 and IR3, and 96.5% between IR2 and IR3. Furthermore, distance tree of blast results represent IR sequences are located with five leaves in a branch.
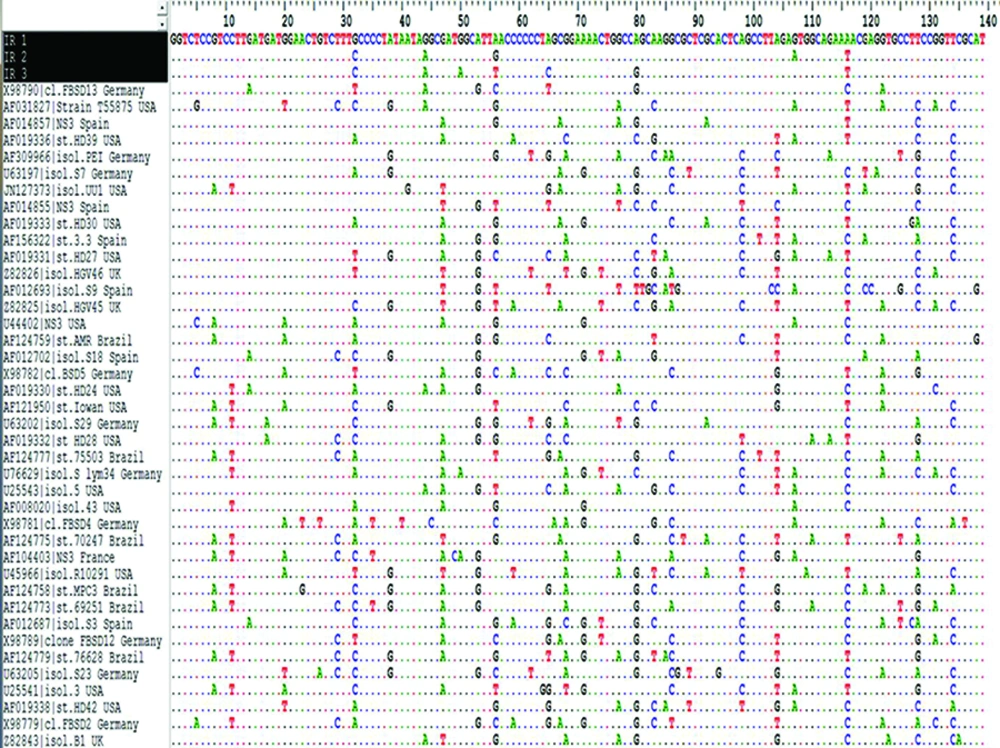
The multiple sequence alignment (MSA) by Bioedit software for Iranian partial NS3across other NS3 genes from different geographical region of world is represented in Figure 2. Results showed highly variation and significant differences in NS3 nucleotide sequences between GBV-C isolates from geographical regions and some little changes in IR sequence. Nucleotide changes were distributed evenly throughout the NS3 nucleotide analyzed fragment. Column of sequences start with three Iranian sequences (with black background) and then followed with others from ascending to descending.
4.3. Phylogenetic Analysis
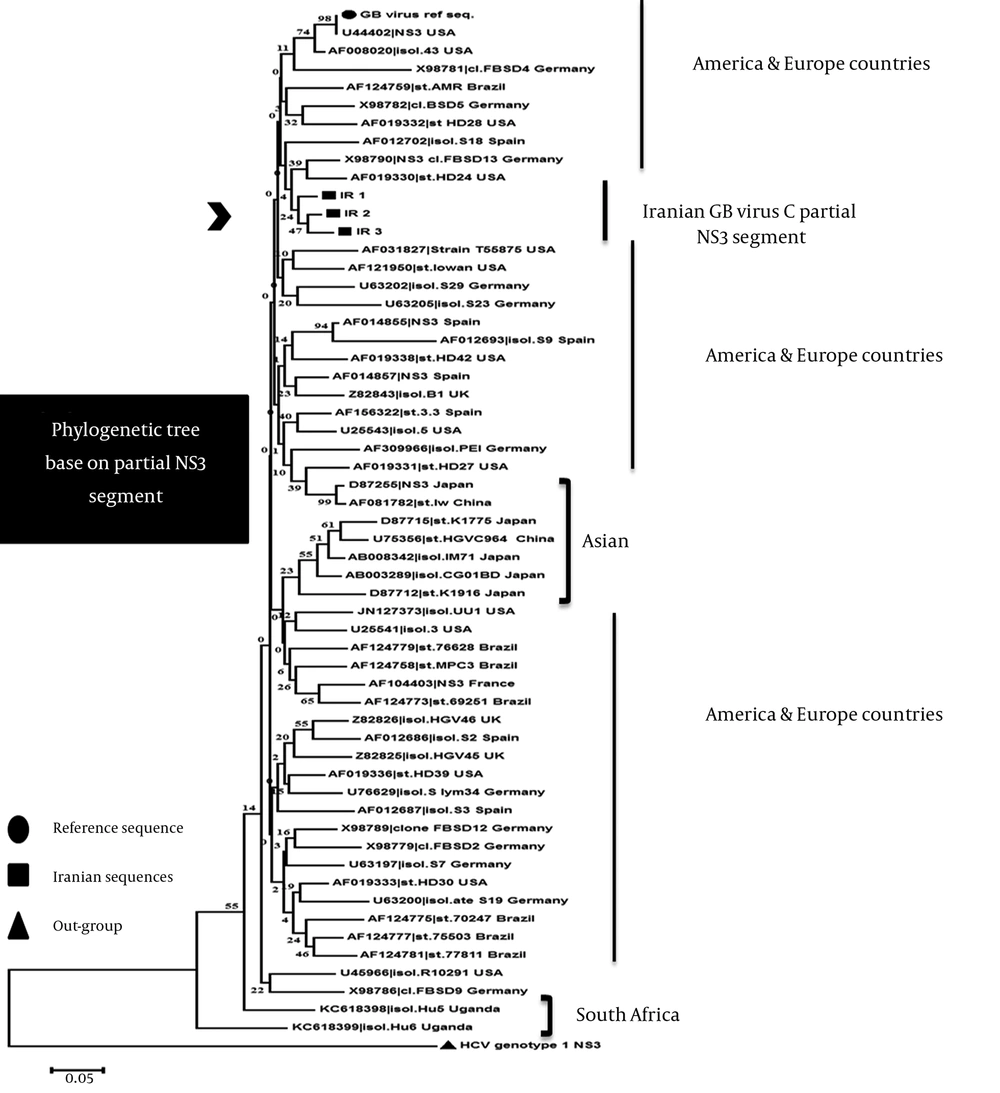
For constructing phylogenetic tree, Iranian partial NS3 gene along with other sequences were constructed on the basis of established genetic relatedness by MEGA 6 software (19) (Figure 3). Phylogenetic tree shows the close relationships between Iranian NS3 sequences and other isolated from different geographical regions of world. IR sequences located in sister branch and most similar sequences in Iran clad are X98790 (Germany), AF019330 (the United States) and then AF012702 (Spain). Moreover, the nearest relatives (nearest clades/group) to IR sequences are AF008020 and U44402 (the United States) reference sequence followed by AF124769 (Brazil), X98782 (Germany), AF019332 (the United States), and X98781 (Germany). Genotype of Iranian isolates is probably similar to European country and US one. The lowest similar sequences were obtained for sequences from South Africa (Uganda) that belong to genotype 5 of GBV-C by reviewing their annotation in NCBI. All sequences from South Africa and South-East Asia, however, are clustered together and are separated from sequences of other isolates collected in Europe and the United States.
Phylogenetic results from Europe, the United States, and Brazil revealed their close relationship and high polymorphism in this gene with regard to different geographical regions. It reflects that they have similar frequent genotypes. Asian sequences (China and Japan) mainly clustered with each other in a clad, except accession numbers D87255 and AF081782, which are more similar to the United States and European countries. One possible explanation for some inconsistent grouping of sequences might be the short coding regions.
| Hits | Maximum Score | Identity, % | Country | Accession Number |
|---|---|---|---|---|
| 1 | 213 | 94 | Spain | AF014857 |
| 2 | 207 | 94 | Germany | X98785 |
| 3 | 207 | 94 | Germany | X98790 |
| 4 | 207 | 94 | USA | U44402 |
| 5 | 202 | 93 | Germany | X98783 |
| 6 | 196 | 92 | Spain | AF014855 |
| 7 | 196 | 92 | The United Kingdom | Z82840 |
| 8 | 196 | 92 | Spain | AF077148 |
| 9 | 196 | 92 | USA | AF019343 |
| 10 | 191 | 91 | Spain | AF014874 |
aRanking of sequences is based on their maximum score and identity which arranged ascending to descending, respectively.
Comparison of Iranian Partial NS3 Gene With the Nucleotide Sequences of Other NS3Genes Obtained From Different Geographical Regions of World . This figure just showed part of the multiple sequence alignment result as produced in Bioedit7.9 software. Conserve, semi-conserved, and variable regions along NS3sequences can be seen. All residues that are identical to the top sequence in an alignment are marked by a dot (.).
Tree was constructed using the neighbor-joining (NJ) algorithm based on differences in NS3sequences of different isolates. Units at the bottom of the tree indicate the number of substitution events. The length of each pair of branches represents the distance between sequence pairs. The dataset was re-sampled 10000 times using the Bootstrap method. The Sequence Information at the tips of the branches includes an accession numbers of the sequences and t name of isolate (isol.), clone (cl.), and strain (str.) for each sequence. Some redundant, very short and incomplete Sequences, or sequences with ambiguous alignment or containing poly-N stretches were excluded after alignment and tree not showing them.
5. Discussion
The studied hepatotropic RNA virus, namely, GBV-C/HGV, is associated with acute and chronic hepatitis, and eventually may lead to cirrhosis and cancer of liver. The HCV is prevalent in IVDUs and patients with thalassemias, but role of HGV in pathogenesis of hepatitis is not well understood (20).
Infection with HGV is relatively common and has a worldwide distribution (5, 21). For epidemiologic reasons, the common HGV infection in humans, particularly in patients on HD, is of interest for controlling parenterally transmitted viral infection in high-risk patients to control the infection and to estimate co-infection with HBV, HCV, and HIV infections. The prevalence of HGV viremia is common in patients on HD as well as in Europe and Asia (21-23).
In patients infected with HIV-1, HGV RNA-positive ones showed less rapid progression of clinical sign of disease, lower death rates, lower reduction in the number of CD4 T cells, reduced in HIV plasma viral load, and higher quality of life compared with HIV-infected individuals without HGV viremia (24-26).
Until now, six major genotypes were reported in different geographical region of world based on differences between the 5’NCR or E2 sequence (27). The distribution of HGV genotypes varies geographically and data still need to complete. Genotype 1 and 5 are predominantly found in Western and south of Africa, respectively, genotype 2 in Europe and the United States, 3 in Asia, 4 in Southeast Asia, and 6 in Indonesia (27, 28).
Overall, based on phylogenetic analysis and BLAST results of Iranian partial NS3 gene of HGV sampled from patients on HD, there were higher similarity between Iranian and Europe-the United States in HGV NS3 gene. In addition, we recommend a study on HCV-positive patients on HD to compare the patients coinfected with HGV and HCV alone for better understanding the effect of HGV on severity of liver disease.
As previously reported in Iran by PCR-sequencing (29) and PCR-RFLP techniques (30) base on 5’ UTR segment, frequent genotype of this virus in Iranian patients is genotype 2, which is frequent in the United States and Europe. Therefore, this result is consistent with the data reported earlier based on 5’ NTR and revealed genotype 2 as the major genotype of Iranian HGV-positive patient.
One remarkable finding in our study was significant variation between the nucleotide sequences of NS3of HGV although the sequences seemed much more conserved than corresponding HCV sequences (14). The lower than expected variability of HGV may have been indicative of weak selection pressure on these viruses, due to an inadequate immune response. Sequence variation in HGV ranges from 0.5% to 20.7% at the nucleotide level, and the NS3 region of HGV has sequence divergence of 10% to 20% (31).
Regarding homology of Iranian nucleotide sequences, there were 96.4% identity between IR1 and IR2, 94.9% between IR1 and IR3, and 96.5% between IR2 and IR3, suggesting that all included sequences belonged to the same GBV-C/HGV genotype. These results are consistent with previous research in HGV isolates from Spanish (32). Moreover, the homology of NS3 nucleotide sequences of the three Chinese HGV strains were reported to be 92.48%, 89.09%, and 85.34% with GBV-C (U36380), and 89.09%, 85.34%, and 85.34% with HGV (U44402) (33).
Current genotyping approaches, applying for many viruses including HCV, mainly rely on a comparison of partial sequences in specific regions of the viral genome. Attempts to group phylogenetic of GBV-C/HGV isolates based on sequence analysis of subgenomic coding regions need further studies. For instance, comparison of sequences within the NS3 helicase region for isolates failed to differentiate between African, Asian, and European-North American isolates (14, 34).
In conclusion, all probable relationships and variations are not reproduced by analysis of fragments of NS3 coding region, but that may be useful in the development of methods to discriminate different variant groupings of HGV. Furthermore, this study will help to better understanding of genetic characteristics of circulating HGV in Iranian population and its similarities and differences with elsewhere.