1. Background
Currently, an estimation of around 3% of people worldwide are chronically infected with Hepatitis C virus (HCV), which is a global health problem caused cirrhosis and hepatocellular carcinoma (HCC) (1). Throughout the past two decades, HCV was recognized and the subsequent interferon-based therapies were used to treat it. The treatment duration of pegylated interferon plus ribavirin (PEG-IFN/RBV) depends upon HCV genotype for 24 - 48 weeks (2), and the sustained virological response (SVR) has been reported to vary depending on genotype, which is achieved in 40% to 50% of patients infected with genotype 1, in 60% patients with genotype 4, and in 80% or more patients with genotypes 2 and 3 (3). Current chronic hepatitis C (CHC) therapy includes direct antiviral agents (DAAs) to further improve the rates of SVR. Several host factors, such as age, sex, liver fibrosis, HCV viral load and HCV genotype (4) have been reported to affect the efficacy of antiviral therapy. Among which single nucleotide polymorphisms (SNPs) includes rs12979860 and rs8099917 on chromosome 19q13 near IFNL3 (IL28B) gene in genomic region were indicated strongly associating with both spontaneous HCV clearance and response to PEG-IFN-α/RBV treatment by several genome-wide-association studies (GWAS) (5-8). In any event, these factors were not yet adequate to explain the variability in HCV treatment response.
Early in 2013, Prokunina-Olsson and colleagues (9) discovered a novel IFN lambda gene upstream of IFNL3 which was designated interferon lambda 4 (IFNL4). The variant polymorphism (rs368234815, originally designated as ss469415590, ΔG/TT) creates the open reading frame and encodes an IFNL4 protein. Transient over-expression of IFNL4 in a hepatoma cell line can induce STAT1/STAT2 phosphorylation and expression of interferon-stimulated genes. Anyhow, rs368234815 sounds to be a splendid predictor of SVR even better than the traditional rs12979860 (10, 11). In addition, the predictive value depends on HCV genotypes and ethnicity (9, 12). However, the influence of IFNL4 variation on treatment outcome was still less; indeed, studies of IFNL4 in predicting SVR depended on HCV genotypes, ethnicity or treatment regimen included only small numbers of patients and produced controversial data.
To our knowledge, there is lack of meta-analyses to date about the association between IFNL4 (rs368234815) polymorphism and SVR. For that reason, we performed a meta-analysis to overcome the limitations of individual study and to understand the real situation.
2. Evidence Acquisition
2.1. Search Strategy
Relevant studies were identified by a PubMed, Medline, Embase, EBSCO and Web of Science literature search with the following terms: (hepatitis C or hepatitis C virus or HCV or chronic hepatitis C or CHC) and (IFNL4 protein, human or IFNL4 or Interferon-λ 4 or Interferon lambda 4 or ss469415590 or rs368234815). No language or time restrictions were applied and the initial search was done on July 26, 2015. We carefully screened the title and abstract of each publication to select the candidate studies. When articles fulfilled the inclusion criteria, we reviewed the full text. Reference lists of the identified articles were also examined. All abstracts presented at congresses obtaining IFNL4 rs368234815 polymorphism and treatment response were also extensively screened. When discrepancies occurred between reviewers (Qin Wu and Cong Wang), discussion and rereview of the studies were applied and a third reviewer (Xue Zhong Lei) participated to perform a final judgment.
2.2. Study Selection
Inclusion and exclusion criteria were established to categorize the studies prior to initiation of the literature search.
Inclusion criteria were as follows: (1) patients infected with HCV and received treatment (IFN and ribavirin or DAAs combination therapy); (2) treatment response defined as sustained virological response (SVR), i.e. extinction of HCV RNA six months after stopping treatment, or non-SVR, detectable HCV RNA at the end of follow-up; (3) comparing virological response rates in patients with favorable IFNL4 genotype for SVR, defined as rs368234815 TT/TT genotype, and in patients with poor-response IFNL4 genotype for SVR defined as the rs368234815 TT/ΔG genotype or the ΔG/ΔG genotype or TT/ΔG + ΔG/ΔG genotype; have been published as a whole-length article or presented as an abstract at congresses.
Exclusion criteria were as follows: (1) coinfection with hepatitis B virus (HBV) and (or) HIV; (2) decompensated liver cirrhosis or HCC; (3) patients received liver transplant; (4) lacking information on SVR rates or IFNL4 polymorphism, not available comparison of SVR rates between IFNL4 genotype (with and without favorable genotype); (5) studies of reviews, letters, editorials, or clinical cases.
2.3. Data Extraction and Quality Assessment
Data abstraction was systematically performed by two independent investigators (Qin Wu and Cong Wang), the cohort studies were assessed using the standardized data collection forms modified from the Newcastle-Ottawa scale (NOS) (maximum score of 9) (13). Also, the randomized controlled trials (RCTs) were assessed using the Cochrane collaboration’s tool (domain-based evaluation) (14). Discrepancies on scores or any disagreements were solved by discussion. When several publications studying the same cohort existed, only the most extensive data were taken into account. When a research contained several subgroups and some of them did not conform to the inclusion criteria, only eligible subgroups were incorporated into the meta-analysis. When the data were not explicitly reported, we contacted individual studies’ authors to request the data.
2.4. Statistical Analysis
In the meta-analysis, the association between IFNL4 polymorphism and SVR in HCV infected patients was obtained by pooling odds ratio (OR) and 95% confidence intervals (CI). The significance of the pooled OR was calculated by the Z-test, and a P value of less than 0.05 was considered significant. The random-effects model (DerSimonian and Laird method (15)) and fixed effect model (Mantel and Haenszel method (16)) were applied for dichotomous outcomes. We assessed study heterogeneity basing on the Cochran’s Q statistic and I2 statistic. Heterogeneity was considered significant when a Q statistic P < 0.1 or I2 > 50%. However, the results were pooled by the random-effects model when P < 0.10, and the fixed-effects model was used when P > 0.10. In addition, the Galbraith plot was used to detect potential outliers of the heterogeneity (17). This graphical method enabled us to check those studies which could have biased the combined estimate. By the way, we conducted the sensitivity analysis to evaluate the consistency of results and to precisely investigate the heterogeneity. For rs368234815, a recessive model was applied (TT/TT vs. TT/ΔG + ΔG/ΔG). Separate analyses were performed to reduce the potential heterogeneity. Therefore, summarized ORs were stratified by HCV genotypes, ethnicity or treatment regimen. Publication bias was assessed by Begg’s funnel plot and the Egger’s linear regression test (17).
All analyses were performed using Stata software (v. 12.0; Stata, College Station, TX), and a two tailed P value less than 0.05 was considered as a statistical significance.
3. Results
3.1. Search Results
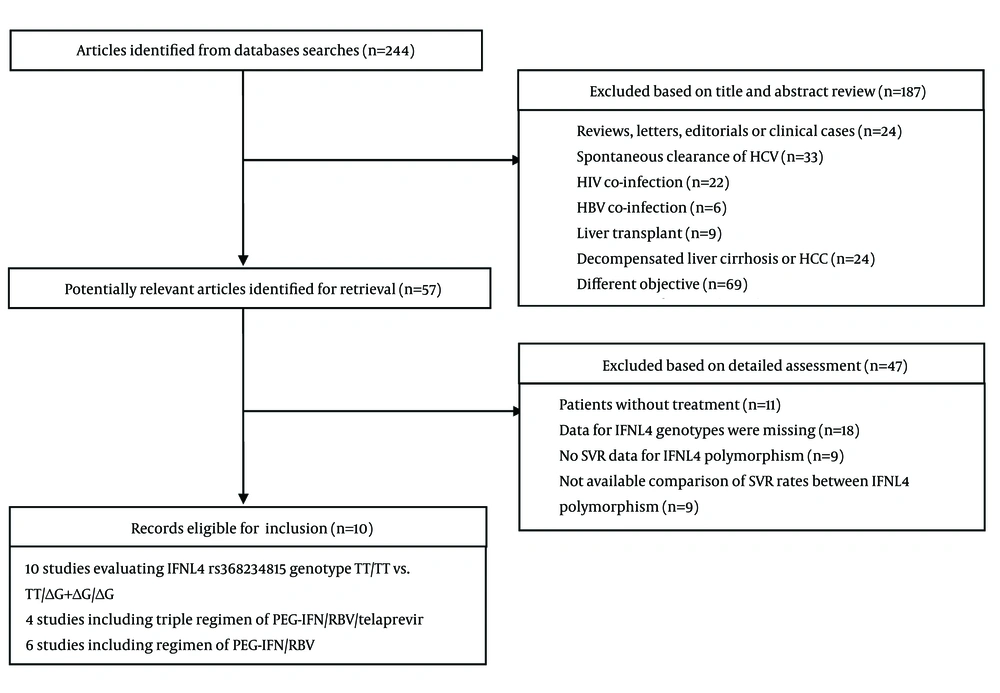
The search strategy yielded 244 entries (Figure 1). Fifty-seven of those 244 were considered to have latent value, and the full texts were gone through thoroughly for detailed evaluation. Several publications studied the same cohort (O’Brien et al. (18) and Prokunina-Olsson et al. (9), Fujino et al. (19) and Nagaoki et al. (20)), we took the most extensive and available data into account. After exclusion according to the inclusion and exclusion criteria, 10 studies involving 4765 patients were eligible for inclusion. According to quality assessment, these studies were relatively well-designed, and the quality assessment of both cohort and RCT studies were at high level. In the 10 studies, patients were treated with PEG-IFN α 2a or 2b and ribavirin with/without telaprevir (TVR). Since two studies contained various ethnicities (21, 22), it was unavailable to compare SVR rates between IFNL4 genotype according to Ethnicity. We incorporated other minority into the Caucasian group in the analysis. The publication year of the studies ranged from 2013 to 2015. The main characteristics of these studies are presented in Table 1.
| First Author/Subgroup ID | Year | Ethnicity (region) | Design | Genotype method | Treatment Trait | Treatment Regime | HCV Genotype | Age (Mean) | Gender ( % for male) | SVR, N | Total, N |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Prokunina-Olsson (9) | 2013 | cohort | TaqMan SNP assay | PEG-IFN/RBV | |||||||
| 1 | African (American) | Naive (Virahep-C) | 1 | 49.1 (45.6 - 52.8) | 110 (65.1) | 48 | 169 | ||||
| N-SVR (HALT-C) | 1 | 51 (46.5 - 56.0) | 82 (56.9) | 10 | 144 | ||||||
| 2 | Caucasian (American) | Naive (Virahep-C) | 1 | 48.1 (42.9 - 52.4) | 120 (65.9) | 94 | 182 | ||||
| N-SVR (HALT-C) | 1 | 49 (45.0 - 53.0) | 559 (75.4) | 133 | 741 | ||||||
| Miyamura (23) | 2014 | Asian (Japan) | RCT | TaqMan SNP assay | Naive, relapse and NR | ||||||
| 3 | PEG-IFN/RBV | 1 | 55.7 ± 11.2 | 71 (50.4) | 70 | 141 | |||||
| 4 | PEG-IFN/RBV/telaprevir | 1 | 58.2 ± 8.7 | 29 (74.4) | 30 | 37 | |||||
| Palmieri (24) | 2014 | Caucasian (Italy | cohort | Real-time PCR | naive | PEG-IFN/RBV | |||||
| 5 | 1 | 55 ± 12 | 310 (58) | 204 | 539 | ||||||
| Susser (25) | 2014 | Caucasian (Germany) | RCT and cohort | TaqMan SNP assay | Naive and ND | ||||||
| 6 | PEG-IFN/RBV | 1 | NA | NA | 194 | 349 | |||||
| 7 | PEG-IFN/RBV/telaprevir | 1 | NA | NA | 52 | 75 | |||||
| 8 | PEG-IFN/RBV | 2 | NA | NA | 51 | 58 | |||||
| 9 | PEG-IFN/RBV | 3 | NA | NA | 105 | 140 | |||||
| 10 | PEG-IFN/RBV | 4 | NA | NA | 135 | 206 | |||||
| Akkarathamrongsin (12) | 2014 | Asian (Thailand) | cohort | Real-time PCR | ND | PEG-IFN/RBV | |||||
| 11 | 1 | 49.0 ± 10.6 | 43 (62.3) | 47 | 69 | ||||||
| 12 | 3 | 49.5 ± 10.5 | 71 (62.3) | 97 | 114 | ||||||
| Covolo (26) | 2014 | Caucasian (Italy) | cohort | TaqMan SNP assay | naive | PEG-IFN/RBV | |||||
| 13 | 1 | 46 ± 11 | 78 (64) | 65 | 120 | ||||||
| 14 | 2, 3 | 46 ± 11 | 88 (55) | 144 | 159 | ||||||
| Stattermayer (21) | 2014 | 745 Caucasian, 6 Asian and 3 African (Austria) | cohort | Real-time PCR | naïve | PEG-IFN/RBV | |||||
| 15 | 1 | 45.1 (44.0 - 46.1) | 259 (60.0%) | 250 | 432 | ||||||
| 16 | 2 | 44.7 (39.9 - 49.4) | 12 (52.2) | 160 | 23 | ||||||
| 16 | 3 | 37.9 (36.4 - 39.4) | 113 (61.1) | 160 | 185 | ||||||
| 17 | 4 | 41.7 (40.1 - 43.3) | 100 (87.7) | 65 | 114 | ||||||
| Nagaoki (20) | 2014 | Asian (Japan) | cohort | Invader assay | PEG-IFN/RBV/telaprevir | ||||||
| 18 | Naive, relapse | 1 | 63 (25 - 79) | 143 (50.5) | 162 | 178 | |||||
| 19 | NR | 1 | 63 (25 - 79) | 143 (50.5) | 64 | 105 | |||||
| Holmes (22) | 2015 | cohort | Real-time PCR | naive | PEG-IFN/RBV | ||||||
| 20 | 28 Asian, 113 Caucasian and 2 Others | 1 | 48.4 (38.1 - 53.6) | 88 (62) | 74 | 143 | |||||
| 21 | 3 Asian, 105 Caucasian and 8 Others | 3 | 43.7 (35.6 - 49.7) | 59 (51) | 94 | 116 | |||||
| Akamatsu (27) | 2015 | Asian (Japan) | cohort | Invader assay | ND | PEG-IFN/RBV/telaprevir | |||||
| 22 | 1 | 62 (24 - 79) | 138 (50.5) | 176 | 226 |
3.2. Meta-Analyses Results
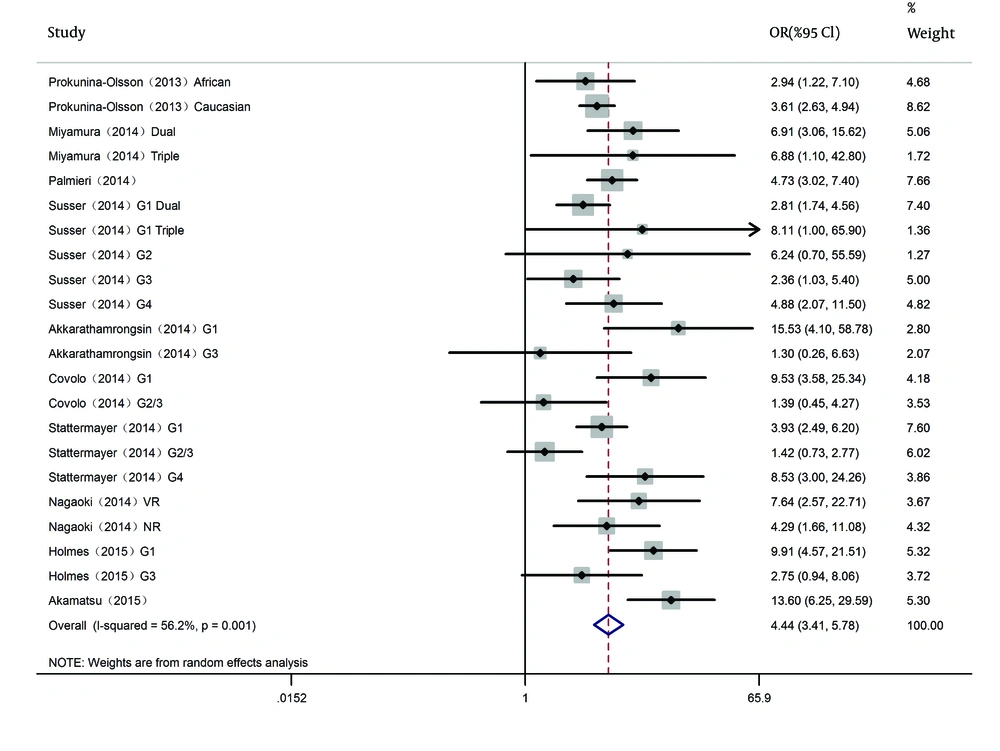
Ten studies stratifying into 22 subgroups were included in the analysis (9, 12, 20-27) (Table 1). A total of 1759 patients had the rs368234815 TT/TT genotype, whereas 3006 patients had the TT/ΔG + ΔG/ΔG genotype. Among those studies, 14 subgroups were on patients with HCV genotype 1 (G1), six subgroups were on patients with HCV genotype 2, 3 (G2/3) and two subgroups were on patients with HCV genotype 4 (G4). In order to reduce the heterogeneity, we separately analyzed studies on the HCV G1, the HCV G2/3 and the HCV G4 patients.
The SVR rate was 45.9% (1674/3650) in G1 patients, 81.9% (651/795) in G2/3 patients and 62.5% (200/320) in G4 patients. For IFNL4 rs368234815, the between-study heterogeneity was significant when all 10 studies (22 subgroups) were pooled for analysis (I2 = 56.2%, P < 0.001); thus, we selected the random-effects model. DerSimonian and Laird method was used (Figure 2). The pooled results presented IFNL4 rs368234815 genotype TT/TT had a higher SVR rate in HCV patients with all HCV genotypes (OR = 4.439, 95% CI: 3.410 - 5.778, P < 0.001).
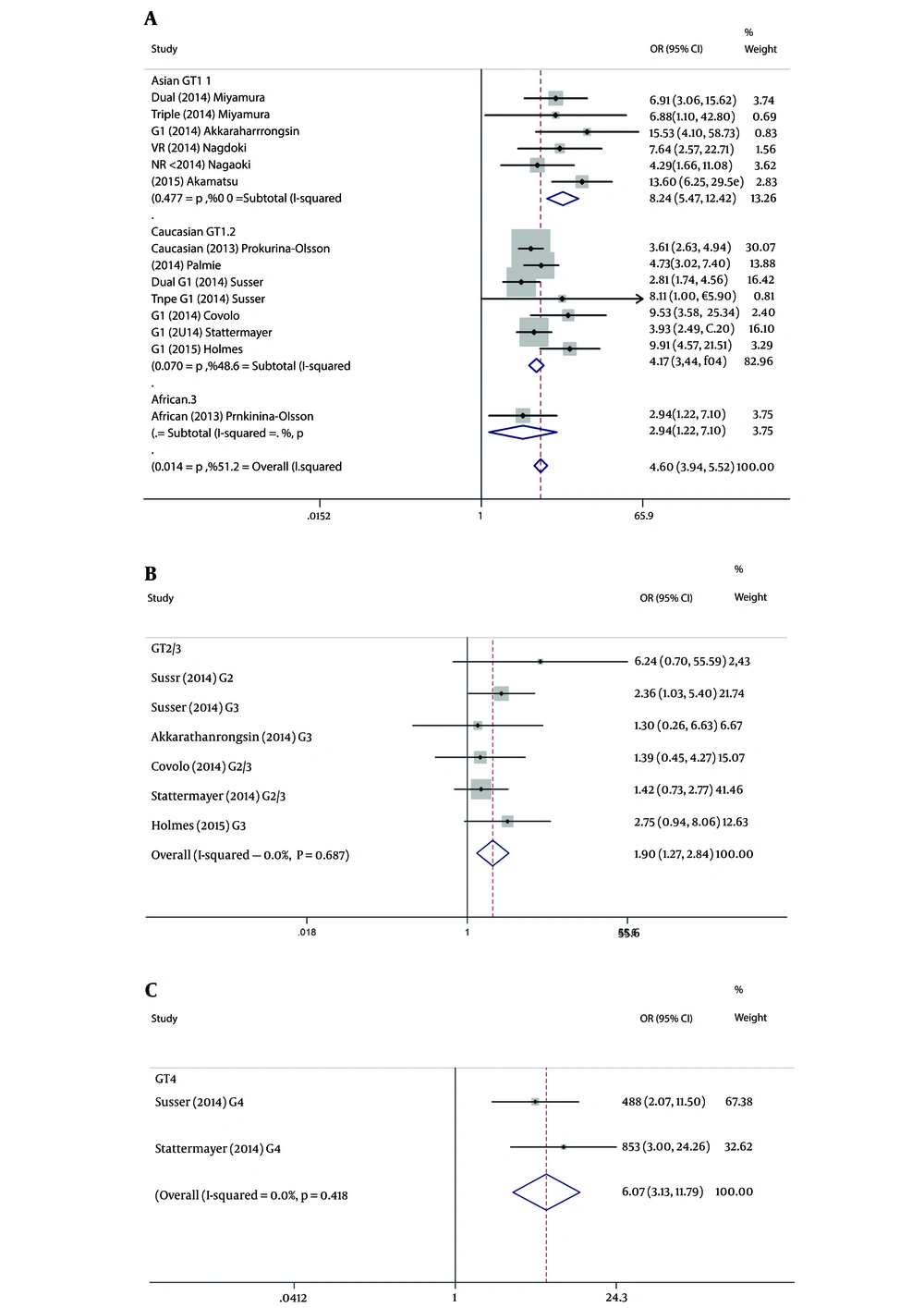
Ten studies including 14 subgroups evaluated the association between sustained virological response and SNP of rs368234815 in HCV G1 patients. When combined data from Asian, the TT/TT genotype showed a rewarding effect compared to ΔG allele, consisted of TT/ΔG + ΔG/ΔG (SVR: 86.4% vs 47.2%; OR = 8.245, 95% CI: 5.475 - 12.416; P < 0.001). When combined data from Caucasian, the TT/TT genotype was also observed to have favorable effect (SVR: 63.9% vs 31.9%; OR = 4.166, 95% CI: 3.441 - 5.042; P < 0.001). Only one study referred to African, TT/TT genotype also had a higher SVR rate (SVR: 37.5% vs 17.0%; P = 0.017). The pooled results from all studies indicated that rs368234815 TT/TT was associated with an increasing SVR rates in HCV genotype 1 patients with Peg IFN/ RBV/TVR or PEG-IFN/ RBV treatment (OR = 4.661, 95% CI: 3.937 - 5.518; P < 0.001) (Figure 3A).
For patients of HCV G2/3, five studies including 6 subgroups evaluated the association between SVR of combined therapy and SNP of rs368234815. Since the G2/3 subgroups were primarily composed of Caucasians (only one among 6 subgroups was Asian), and the between-study heterogeneity was not significant (I2 = 0, P = 0.687), ethnicity stratification analyses were not established. In genotype 2/3 stratification analysis, however, rs368234815 genotype TT/TT was also related to SVR in HCV patients (SVR: 86.4% vs 77.6%; OR=1.896, 95% CI: 1.265 - 2.841; P = 0.002) (Figure 3B).
For patients of HCV G4, similar results were observed from 2 studies including 2 subgroups which were composed of Caucasians. G4 infected patients with rs368234815 TT/TT genotype would achieve higher SVR. Meanwhile, the favorable TT/TT genotype would associate with 6.074-fold probability achieving SVR versus the unfavorable genotype (SVR: 86.8% vs 52.8%; OR = 6.074; 95% CI: 3.129 - 11.788; P < 0.001) (Figure 3C).
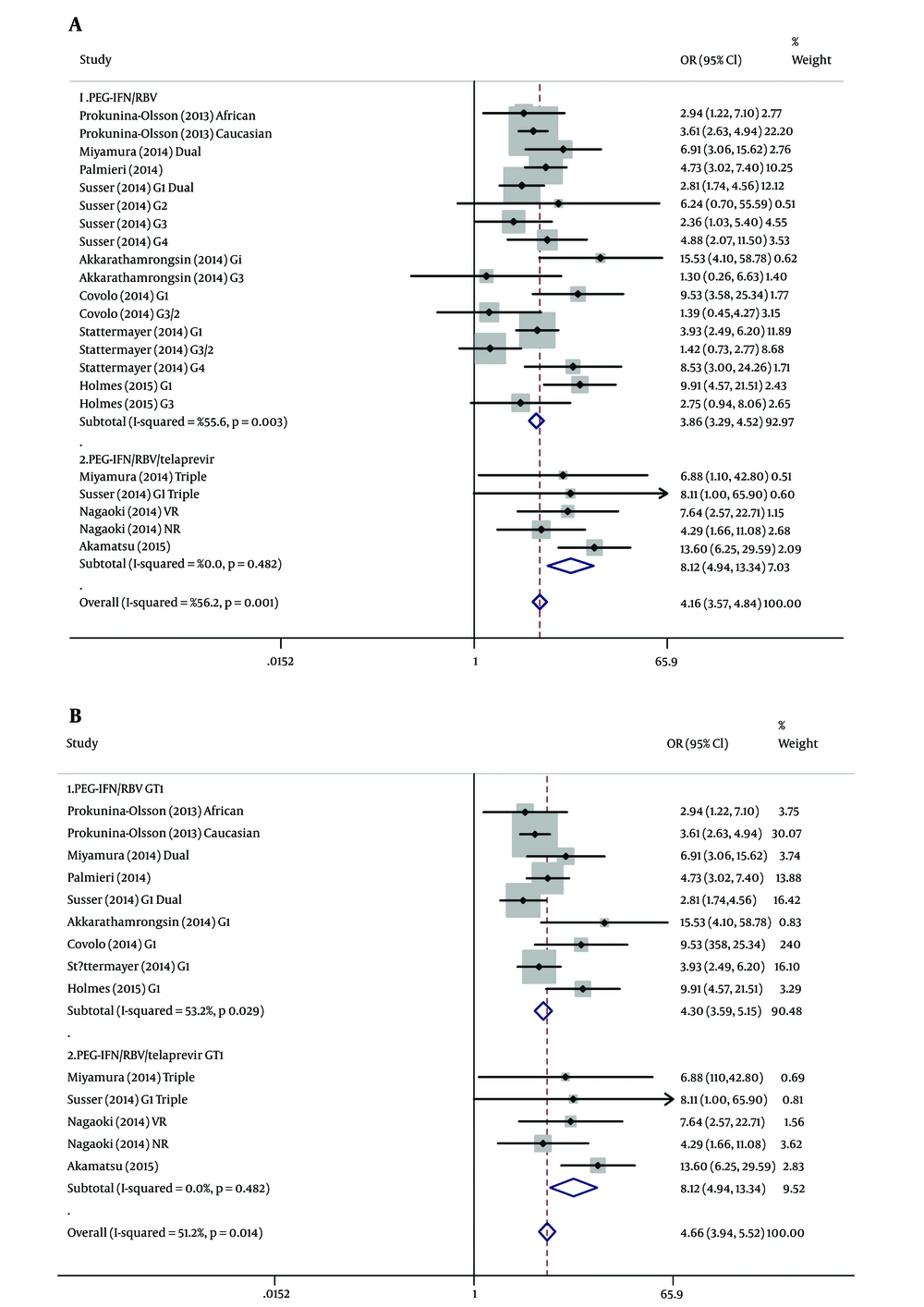
Direct antiviral agents (DAAs) as selective inhibitors of HCV nonstructural 3/4A protease activity have remarkably improved SVR rates in HCV patients (28, 29). Among 10 incorporate studies of this meta-analysis, four of them including 5 subgroups were treated with Telaprevir (TVR), PEG-IFN plus RBV. The SVR rate of patients with triple therapy was significantly higher than patients with dual therapy (77.9% vs. 49.3%, P < 0.001). Overall studies of therapy stratification analyses with the random-effects model were shown in Figure 4A. For patients with dual treatment, the rs368234815 TT/TT genotype showed a higher SVR compared to the TT/ΔG + ΔG/ΔG genotype (OR = 3.857; 95% CI: 3.288 - 4.524; P < 0.001). For patients with triple treatment, similar results were observed (OR = 8.119; 95% CI: 4.942 - 13.340; P < 0.001). Therapy stratification analyses of G1 patients revealed that rs368234815 TT/TT was associated with an increasing SVR rates in HCV patients with PEG-IFN/ RBV treatment (SVR: 63.8% vs 28.6%; OR = 4.298, 95% CI: 3.588 - 5.147; P < 0.001). Meanwhile, the favorable TT/TT genotype would associate with 8.119-fold probability achieving SVR in G1 patients with Peg IFN/ RBV/TVR treatment (SVR: 92.8% vs 57.3%; OR = 8.119; 95% CI: 4.942 - 13.340; P < 0.001) (Figure 4B).
3.3. Heterogeneity Analysis
For IFNL4 rs368234815, the between-study heterogeneity was significant for the pooled analysis (n = 10, subgroups = 22), as well as the virus genotype stratification analysis of G1 (P = 0.014, I2 = 51.2%). For G1 patients, the between-study heterogeneity was only significant for ethnicity stratification analysis of Caucasians (P = 0.070, I2 = 48.6%). In contrast, heterogeneity was not significant for G1 data from Asian (P = 0.477, I2 = 0) (Figure 3A). In addition, the between-study heterogeneity was not significant for any of the other HCV genotypes and ethnicity stratification analysis (Figure 3B and C).
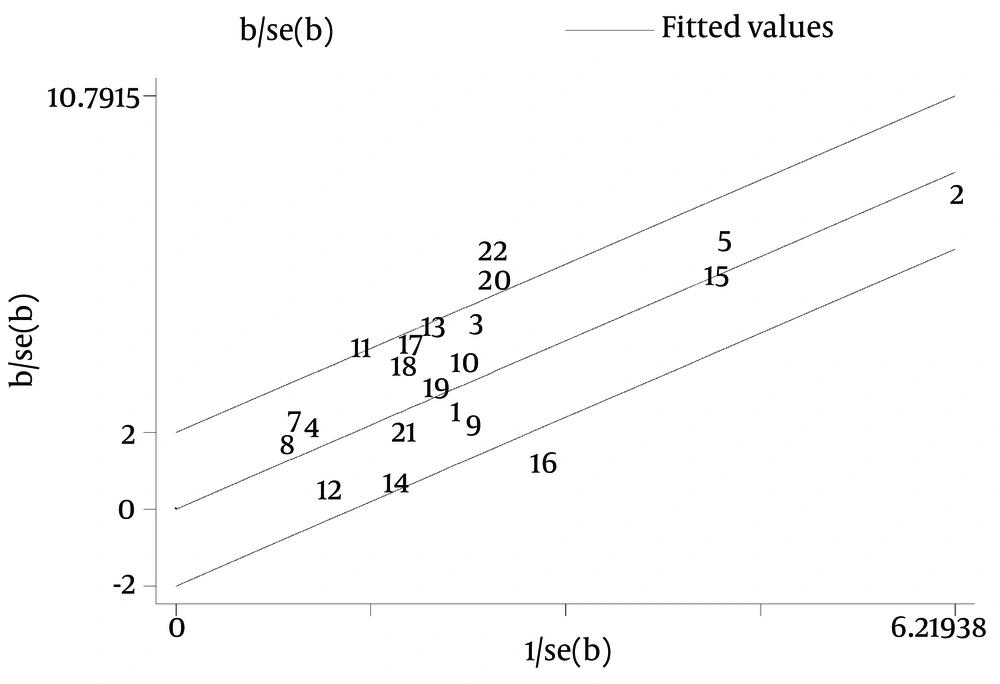
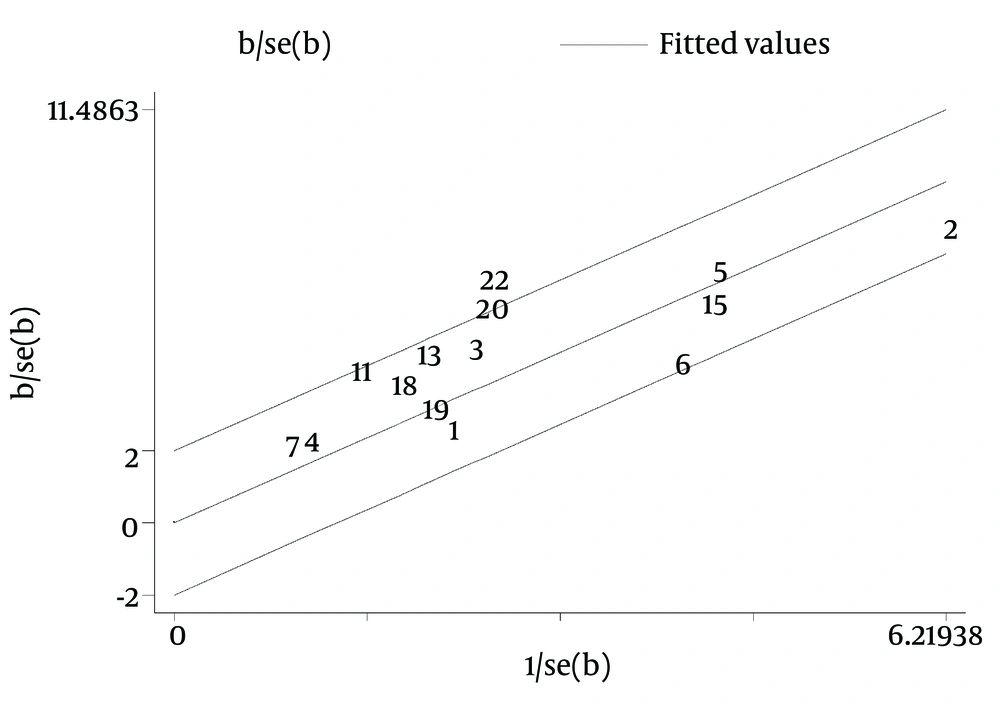
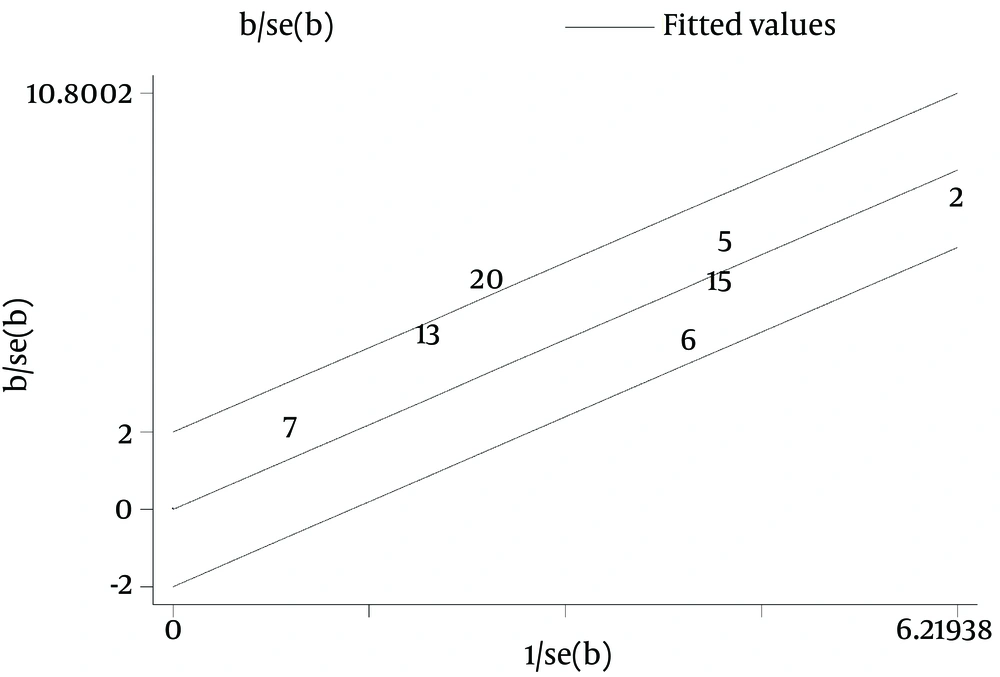
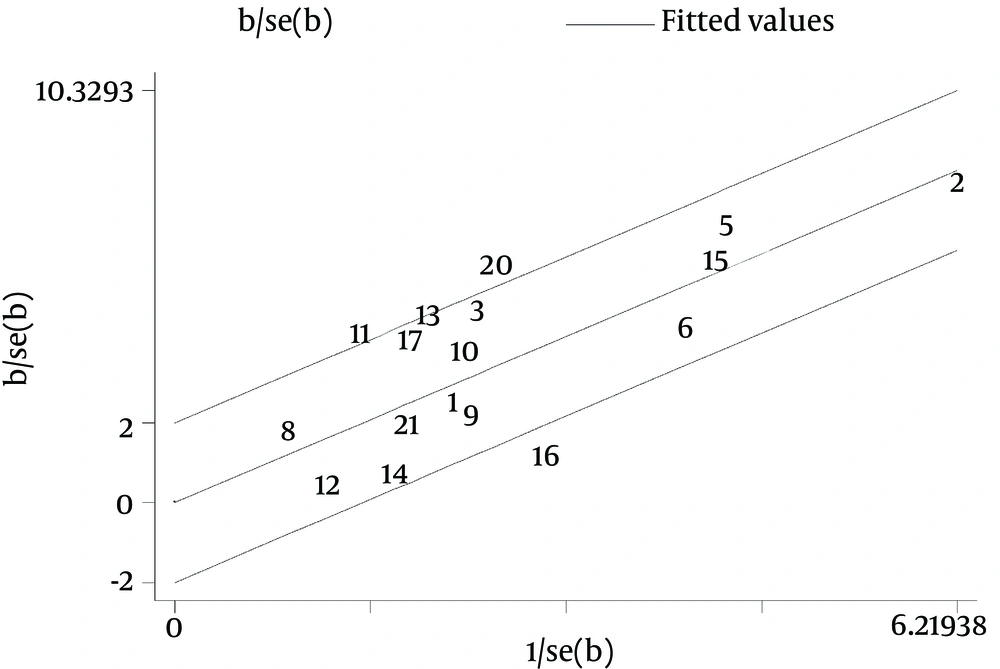
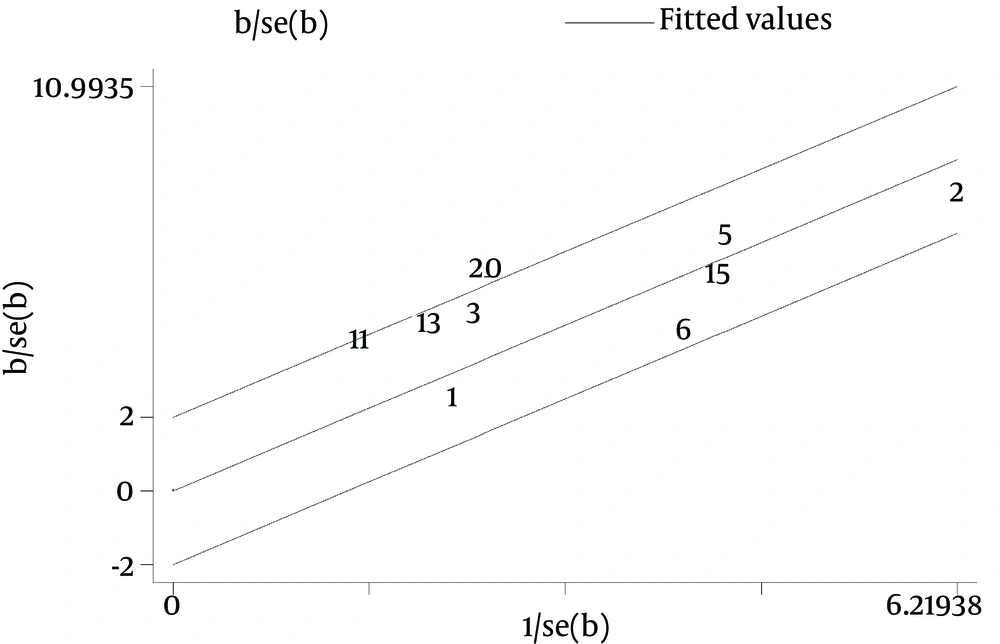
Galbraith plot of the overall pooled studies (n = 10, subgroups = 22) showed Stattermayer’s study on G2/3 subgroup (21), Holmes’s study on G1 subgroup (22) and Akamatsu’s study (27) were outliers as potential sources of heterogeneity (Figure 5). In genotype stratification analyses, Galbraith plot of G1 showed one outlier (27) (Figure 6), while ethnicity stratification analyses plot of G1 Caucasian patients showed one study (22) (Figure 7). In therapy stratification analyses, Galbraith plot of dual treatment showed two outliers (21, 22) (Figure 8), while genotype stratification analyses plot of dual treatment G1 patients showed one study (22) (Figure 9).
Therefore, we excluded the tree heterogeneity subgroups adjusted and reduced the heterogeneity for the overall studies (P = 0.154, I2 = 25.1%), furthermore excluded the one source from the G1 analysis (P = 0.104, I2 = 36.0%), as once more excluded the one outlier from the Caucasian G1 analysis (P = 0.270, I2 = 21.7%). In therapy stratification analyses, we studied on patients with dual therapy and excluded the two heterogeneity subgroups (P = 0.108, I2 = 35.7%); on G1 dual therapy studies, we excluded the one outlier (P = 0.94, I2 = 42.6%). We reappraised the correlation of rs368234815 genotype TT/ TT with SVR in overall studies, in G1 patients, only with the values of ORs slightly altered and still found statistically significant results (data not shown).
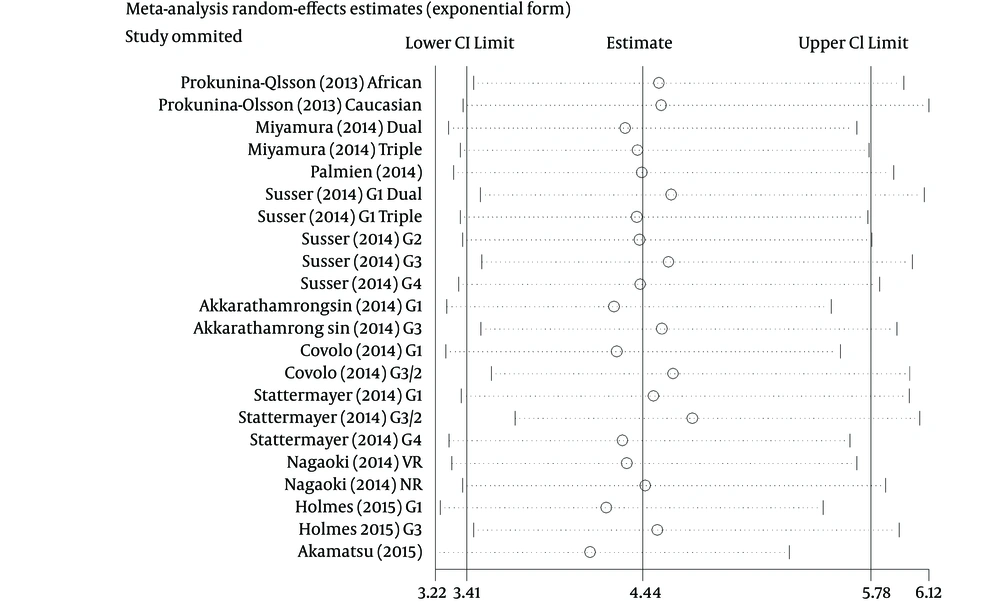
3.4. Sensitivity Analysis
Sensitivity analysis was performed by sequential omission of every study respectively. Results showed that the omission of any study made no significant difference; thus, our results were reliable (Figure 10).
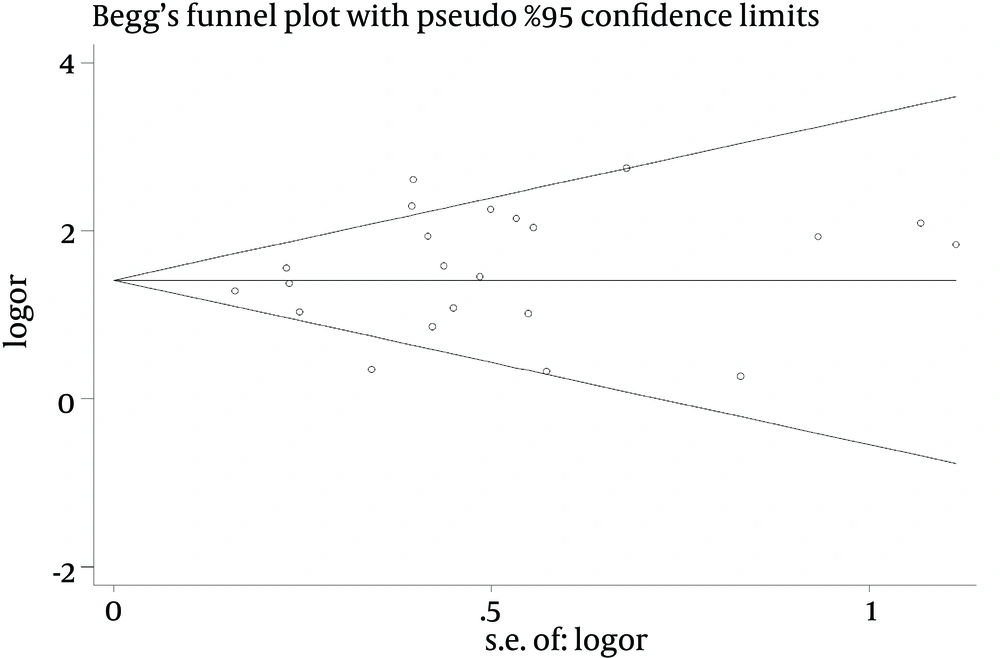
3.5. Publication Bias
Publication bias was not observed in studies using Begg’s test and Eggers’ test respectively in this meta-analysis, as shown in Table 2 (Figure 11).
| Group | P for Begg | P for Egger |
|---|---|---|
| Overall studies | 0.735 | 0.294 |
| HCV GT 1 | 0.274 | 0.058 |
| HCV GT 1 Asian patients | 1.000 | 0.936 |
| HCV GT 1 Caucasian patients | 0.548 | 0.099 |
| HCV GT 2/3 | 0.260 | 0.358 |
| HCV GT 4 | NA | 1.000 |
| Patients with dual treatment | 0.967 | 0.681 |
| Patients with triple treatment | 1.000 | 0.644 |
4. Conclusions
To our knowledge, there is lack of study collected and summarized the relationship between IFNL4 rs368234815 and treatment response (SVR) in chronic HCV patients. Since host genetic polymorphisms near the interferon-λ-3 (IFNL3, formerly known as IL28B) gene was recognized to be strongly associated with response to PEG-IFN/RBV therapy for chronic HCV, several clinical trials have revealed a prediction of treatment outcome by genotyping for the IL28B SNP (30-32). One of our previous studies also found that rates of SVR was significantly higher, and nonresponse and relapse rates were significantly lower in patients, who carried the favorable IFNL3 genotype (C/C for rs12979860) compared to the adverse IFNL3 genotypes (C/T and T/T) (33). However, the exact mechanisms behind this association were still only partially understood, and GWAS of IFNL3 SNPs were not reported to be functional.
In 2013, Prokunina-Olsson et al. pinpointed a functional dinucleotide variant (rs368234815, TT and ΔG alleles) between IFNL3 and IFNL2, leading to explain the association between IFNL3 genotype and SVR. The ΔG allele led to expressing a novel protein, interferon-λ-4, which encoded by the IFNL4 gene. Expression of interferon-stimulated genes (ISGs) could be induced by overexpression of IFN-L4 in a hepatoma cell line (9). In one study, patients with IFN-α–free DAA therapy included sofosbuvir along with RBV, carrying the IFNL4-ΔG had a slower early viral decay and decreased drug efficacy (34). IFNL4 rs368234815 polymorphism has also been an important predictor of response liver transplantation (35).
Our meta-analysis included 10 studies involving 4765 patients, evaluated the prediction value of the new SNP rs368234815 on treatment outcome to dual- and triple-therapy in HCV genotype 1, 2, 3 and 4 infected Asian, Caucasian and African patients. And this meta-analysis illuminated that a 4.439-fold significant increase of the possibility to obtain SVR compared patients with rs368234815 TT/TT genotype to TT/ΔG + ΔG/ΔG genotype, irrespective of HCV genotypes, ethnicity or treatment regimen. The SVR rates have been reported to vary depending on HCV genotype (2, 3) and the duration of therapy (36, 37). The heterogeneity in response to IFN-based treatment existed among different ethnic or racial groups worldwide (38). Meanwhile, the HCV treatment regimen underwent a major change after the approval of direct antiviral agents (DAAs). Therefore, separate analyses on HCV genotypes, ethnicity and treatment regimen were performed to reduce the potential heterogeneity.
Genotype stratification analyses revealed that rs368234815 TT/ TT was associated with higher SVR in G1, G2/3 and G4 HCV patients. Ethnicity stratification analyses of G1 patients showed that rs368234815 TT/TT was associated with SVR in Asians, Caucasians and Africans. In patients with PEG-IFN/RBV or PEG-IFN/RBV/TVR treatment, the SVR rate of patients with triple therapy was significantly higher than patients with dual therapy. The tendency of IFNL4 rs368234815 predicted value in different treatment regime was similar. Therapy stratification analyses performed a higher SVR rate in rs368234815 TT/ TT genotype compared to the TT/ΔG + ΔG/ΔG genotype both in patients with dual therapy or triple therapy. Thus, detects for IFNL4 rs368234815 polymorphism may be beneficial to guide the clinician in the individualization of treatment regimen and design.
Apart from what mentioned above, some possible limitations should be taken into account when interpreting the conclusions of our meta-analysis. Firstly, meta-analysis reflects the methodological problems of the included studies. Secondly, therapeutic effect would be interpreted prudently for multiple influence variables known or unknown, such as patients with unsuccessful or unknown treatment history, may reversely correlate with treatment effect of chronic HCV patients (39). In this analysis, we could not adjust this potential factor because very limited data were reported on it and not available to compare SVR rates between IFNL4 genotype stratified by treatment history (9, 12, 20, 23-25, 27). Thirdly, the sample size of the genotype 2/3 and genotype 4 groups were relatively small. Given the insufficient studies on genotype 4 patients, we did not conduct ethnicity stratification analysis. This might be accomplished when future studies incorporating larger groups of genotype 2, 3, 4 patients and a multitude of studies investigating different ethnicities to validate the associations with IFNL4 polymorphism. Finally, meta-analyses are most persuasive when implemented with all available data, including those have not been published. When the data were not explicitly reported, we tried to contact some authors of individual studies to request the data, but failed. Therefore, although Begg’s test and Egger’s test of publication bias turned out to be acceptable which meant no such bias existed, the publication bias still may have occurred. However, sensitivity analysis consolidated our results and the potential publication bias might not influence the credibility of this meta-analysis.
In conclusion, this meta-analysis illuminates that rs368234815 TT/ TT genotype is a strong predictor of SVR in HCV patients, irrespective of HCV genotypes, ethnicity or treatment regimen. Thus, detects for IFNL4 rs368234815 polymorphism may be beneficial to guide the clinician in the individualization of therapy and design.