1. Background
One of the most important members of Retroviridae family is a lentivirus called human immunodeficiency virus (HIV) that is the cause of acquired immunodeficiency syndrome (AIDS) (1). According to the last report of joint United Nations program on HIV/AIDS (UNAIDS), above 35 million people suffered from HIV in 2013 (2). HIV infects immune system cells and expresses CD4 (cluster of differentiation 4) surface glycoprotein and thereby can lead the immune cells to lose their efficiency (3). Similarly, the hepatitis viruses B and C (HBV and HCV) have a place among top 10 main infectious agents leading to death worldwide (4-6). Due to similar routes of transmission, HBV and HCV are usually transmitted with HIV and the concurrent HIV, HCV, and HBV infections are considered as a major health problem (7, 8). Concomitant infection with these three viruses may lead to higher HCV load, increased risk of hepatocellular carcinoma, and rapid progression to AIDS compared to mono-infection (7). It has been proven that molecular mechanisms of a co-infection situation mainly differ from those of mono-infection condition. With regard to this issue, one of the most important differences is attributed to the alteration of micro ribonucleic acids (miRs) expression due to different virus’s factors or host defense against viruses (9). A miR is a small, non-coding and evolutionary conserved RNA that contains 18 - 25 nucleotides (10). MiRs can interact with various mRNA targets by base pairing in 3′-untranslated region (3′-UTR) and may affect many cellular functions such as cellular growth and differentiation, metabolism, and apoptosis (10). It has been previously shown that some miRs may efficiently affect viral replication (11, 12). Among all micro RNAs, miR-29 has been clearly shown to inhibit HIV replication (13). The miR-29 family consists of three major members: miR-29a, mir-29b, and miR-29c. Bandyopadhyay et al. (14) previously showed that all members of miR-29 are down-regulated in HCV patients. Besides, they also revealed that down-regulation of miR-29 in hepatocytes by HCV infection increases the risk of collagen synthesis in cell culture while induction of miR-29 reverses this trend. In addition, down-regulation of miR-29a and miR-29b has been also shown in HIV mono-infected patients (15). Experimental evidence shows that the main targets of miR-29 are myeloid leukemia cell differentiation protein, T-cell leukemia/lymphoma 1 oncogene, DNA methyltransferases 3A and 3B (DNMT3A and DNMT3B), and Zinc finger protein 36 homolog (16-19). To the best of our knowledge, there are no other studies about the comparison of miR-29 expression in HIV mono-infected patients and HIV/HCV co-infected patients.
2. Objectives
Therefore, the present study aimed to evaluate miR-29a-5p expression in mono- and co-infections as a novel diagnostic marker for screening and monitoring of infected patients. Furthermore, the possible correlation of miR-29a-5p with clinical and laboratory features of infected subjects was studied.
3. Methods
3.1. Subjects
In this case control study, 165 HIV-positive patients were enrolled who had a regular follow-up at the injection drug user center (IDU) in Sanandaj (Kurdistan, Iran) between April 2015 and February 2016. Out of 165 patients, 44 had only HIV infection (mono-infection group) and 121 were diagnosed with both HIV and HCV infections (co-infection group). Additionally, 40 healthy participants were enrolled as a control group. Written informed consent was obtained from participants and the project was approved by the research ethics committee of Kurdistan University of Medical Sciences (Iran) (project and ethic code: MUK.REC.1394/281, February 2016). Demographic, clinical, and laboratory data as well as information on the specific therapy were recorded. Criteria for exclusion were blood CD4+ cell counts below 200 cell/µL and more than 3 years history of HIV infection (20). In addition, patients who had a history of known causes of liver disease, diabetes mellitus, and cardiac or renal failure were excluded from the study. All men had been infected with HIV via multiperson use (sharing) of drug injection equipments, whereas sexual transmission was the main cause of HIV infection in female subjects.
3.2. Blood Sample Collection
Fasting blood samples were collected from HIV patients; sera were separated and stored at 70°C until simultaneous analysis. Aliquots of the EDTA whole blood samples were used for CD4+ cell count by flow cytometry (FACS Counter, Becton and Dickinson, San Jose, CA) and miRs analysis. Serum total Bil and AST, ALT, and ALP activities, as markers of liver injury, were determined by colorimetric methods using commercial enzymatic kits, according to the manufacturer’s instructions (Pars Azmoon, Tehran, Iran). In addition, Fibrosis-4 (FIB-4) index and AST to platelet ratio index (APRI) score were calculated as previously described (21, 22).
3.3. Immunological Test For Confirmation of Disease in the Studied Population
HIV antibodies were first detected in serum samples by an ELISA kit (Dia lab, Austria); positive results were double checked and finally confirmed by Western Blotting (Biorad, Segrate, Italy) (23). In addition, serum samples were tested for HCV infection by anti HCV antibody ELISA kits (Delaware Biotech Inc., USA) and positive results were reconfirmed by HCV recombinant immunoblot assay (RIBA) (Chiron Corp., Emeryville, California) (24, 25).
3.4. Quantitative Real-Time PCR Analysis
Total RNA was extracted from 100 µL whole blood samples using the RNX-Plus reagent according to the manufacturer's instructions (SinaClon, Tehran, Iran).
Qualitative and quantitative assessment of isolated RNA was carried out by electrophoresis and spectrometric methods (26). For miR quantification by Real-Time PCR in all samples, 10 µL of total.
RNA was reverse-transcribed in a 20 µL reaction mix using the BONmiR1st-strand cDNA synthesis kit (stem cell technology research center, Tehran, Iran) following the manufacturer’s recommendations.
Then, cDNA was used in each of the real-time PCR assays with the BONmiR QPCR Kit (stem cell technology research center, Tehran, Iran) on a rotor gene 6000 thermal cycler (Corbett life science) according to the manufacturer’s instructions. Real-time PCR analyses of miR-29a-5p were carried out in triplicate. The miR-29a-5p levels were normalized using Snord47 RNA as reference RNA. In clinical samples, the mean of miR-29a-5p Cts in all the normal samples was determined and compared to the obtained Cts of each patient. The same procedure was performed for Snord47, as internal control gene, in order to normalize the expression data.
3.5. Statistical Analysis
Statistical analysis was performed using SPSS Statistics v. 16.0 for Windows (SPSS, Chicago, IL). One sample Kolmogorov Simonov test was applied to determine normal distribution of data. Data were presented as mean ± standard deviation (SD). Data were analyzed by nonparametric tests including Spearman rank correlation coefficient, Kruskal-Wallis for more than two groups, and Mann-Whitney test for unpaired comparisons. Multivariate analyses for the presence of liver injury or fibrosis were performed using multivariate analysis of variance (MANOVA). A P value below 0.05 was considered statistically significant.
4. Results
A total of 165 patients (137 males and 28 females) with the mean age of 38.3 ± 7.7 years were diagnosed as HIV positive, according to WHO criteria for diagnosis of HIV (27). The control group consisted of 40 healthy individuals with negative immunological tests and the mean age of 34.7 ± 7.03 years. Among the study subjects, 121 were diagnosed with HIV/HCV co-infection (mean age of 38.49 ± 6.88 years). The control group comprised 31 male and 9 female subjects while the corresponding numbers in the HIV/HCV co-infection group were 115 and 6, respectively. The frequency of mycobacterium tuberculosis (MTB) was 2 in HIV mono-infection and 4 in HIV/HCV co-infection group. Out of 165 patients, 72 (43.63%) were under treatment with Pegylated interferon and ribavirin regimens, 22 (13.33%) cases received ART, and 71 people (43.04%) were new cases and hence had not received any antiviral drug. Tables 1 and 2 show the clinical and laboratory features of the study subjects.
| Healthy Controls (N = 40) | HIV Mono-Infection (N = 44) | HIV/HCV Co-Infection (N = 121) | |
|---|---|---|---|
| Age-years, mean (range) | 36.7 (25 - 51) | 37.79 (20 - 62) | 38.49 (20 - 59) |
| Gender (male/female) | 31/9 | 22/22 | 115/6 |
| Receiving medication-number (yes/no) | - | 22/22 | 72/49 |
| Regular drug use-number (yes/no) | - | 17/5 | 42/30 |
| Weight loss in the last two years-number (yes/no) | - | 24/20 | 69/52 |
| Mouth sores-number (yes/no) | - | 5/39 | 23/98 |
| Pruritic skin-number (yes/no) | - | 7/37 | 36/85 |
| Fungal nail infection-number (yes/no) | - | 0/44 | 2/119 |
| Diarrhea-number (yes/no) | - | 21/23 | 63/58 |
| Oral candidiasis-number (yes/no) | - | 3/41 | 7/114 |
| Pneumonia-number (yes/no) | - | 7/37 | 24/97 |
| Persistent fever-number (yes/no) | - | 22/22 | 68/53 |
| Herpes (genital or anorectal)-number (yes/no) | - | 3/41 | 4/117 |
| Healthy Controls (N = 40) | HIV Mono-Infection (N = 44) | HIV/HCV Co-Infection (N = 121) | P Value | |
|---|---|---|---|---|
| AST, U/L | 17.12 ± 4.54 | 30.92 ± 17.89 | 48.2 ± 24.34 | 0.011 |
| ALT, U/L | 14.29 ± 3.69 | 28.29 ± 20.02 | 40.71 ± 23.58 | 0.015 |
| ALP, U/L | 173.71 ± 34.24 | 273.85 ± 111.76 | 259.57 ± 95.05 | 0.001 |
| Total Bil, mg/dL | 0.49 ± 0.09 | 0.54 ± 0.16 | 0.57 ± 0.21 | 0.236 |
| CD4+ count, cell/µL | 1047.94 ± 34.24 | 429.91±239.13 | 418.42 ± 283.33 | < 0.0001 |
| WBC count (× 103 cell/µL) | 6.47 ± 1.16 | 5.83 ± 1.72 | 5.51 ± 1.85 | 0.83 |
| RBC count (× 106 cell/µL) | 5.12 ± 0.61 | 4.82 ± 0.93 | 4.87 ± 0.79 | 0.361 |
| Plt (× 105/µL) | 222.06 ± 52.92 | 238.00 ± 87.29 | 203.66 ± 67.42 | 0.019 |
| RDW-CV (%) | 12.21 ± 3.97 | 13.94 ± 3.17 | 14.33 ± 1.78 | 0.106 |
| MCV, fL/cell | 84.75 ± 8.49 | 94.20 ± 14.61 | 99.57 ± 42.17 | 0.208 |
| MCH, pg/cell | 28.37 ± 3.59 | 28.00 ± 4.68 | 28.18 ± 4.39 | 0.95 |
| MCHC, g/dL | 29.9 ± 2.92 | 29.69 ± 1.11 | 29.13 ± 2.48 | 0.68 |
| Hb, g/dL | 14.92 ± 2.25 | 13.19 ± 1.82 | 13.49 ± 1.8 | 0.361 |
| FIB-4 | 0.69 ± 0.19 | 0.92 ± 0.39 | 1.62 ± 0.32 | 0.001 |
| APRI | 0.27 ± 0.09 | 0.44 ± 0.27 | 0.71 ± 0.48 | 0.001 |
Abbreviations: AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALP, alkaline phosphatase; Bil, bilirubin; WBC, white blood cell; RBC, red blood cell; Plt, platelets;RDW, red cell distribution width; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; Hb, hemoglobin.
aData represented as mean ± SD.
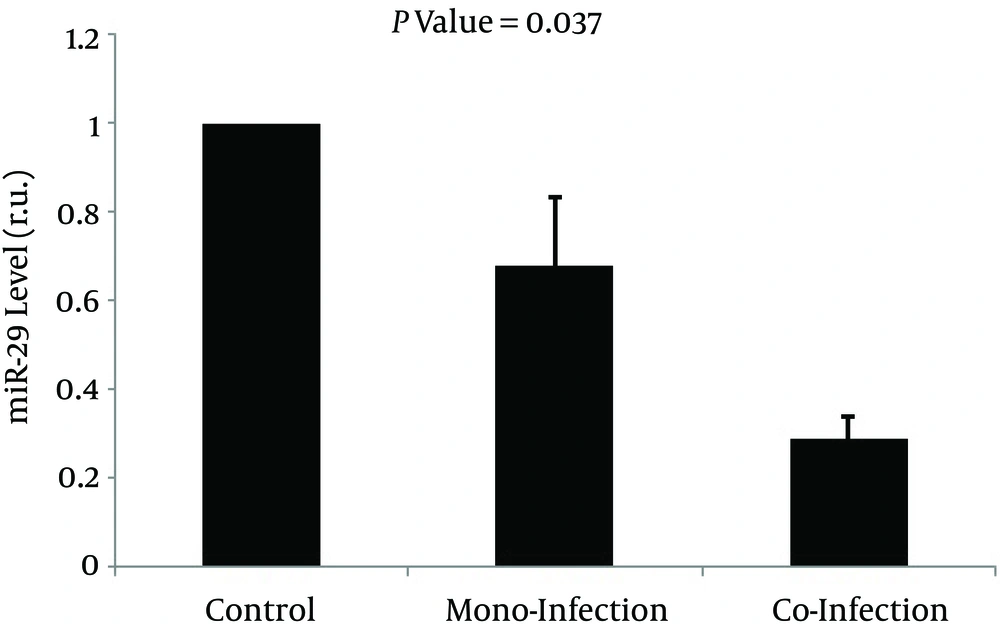
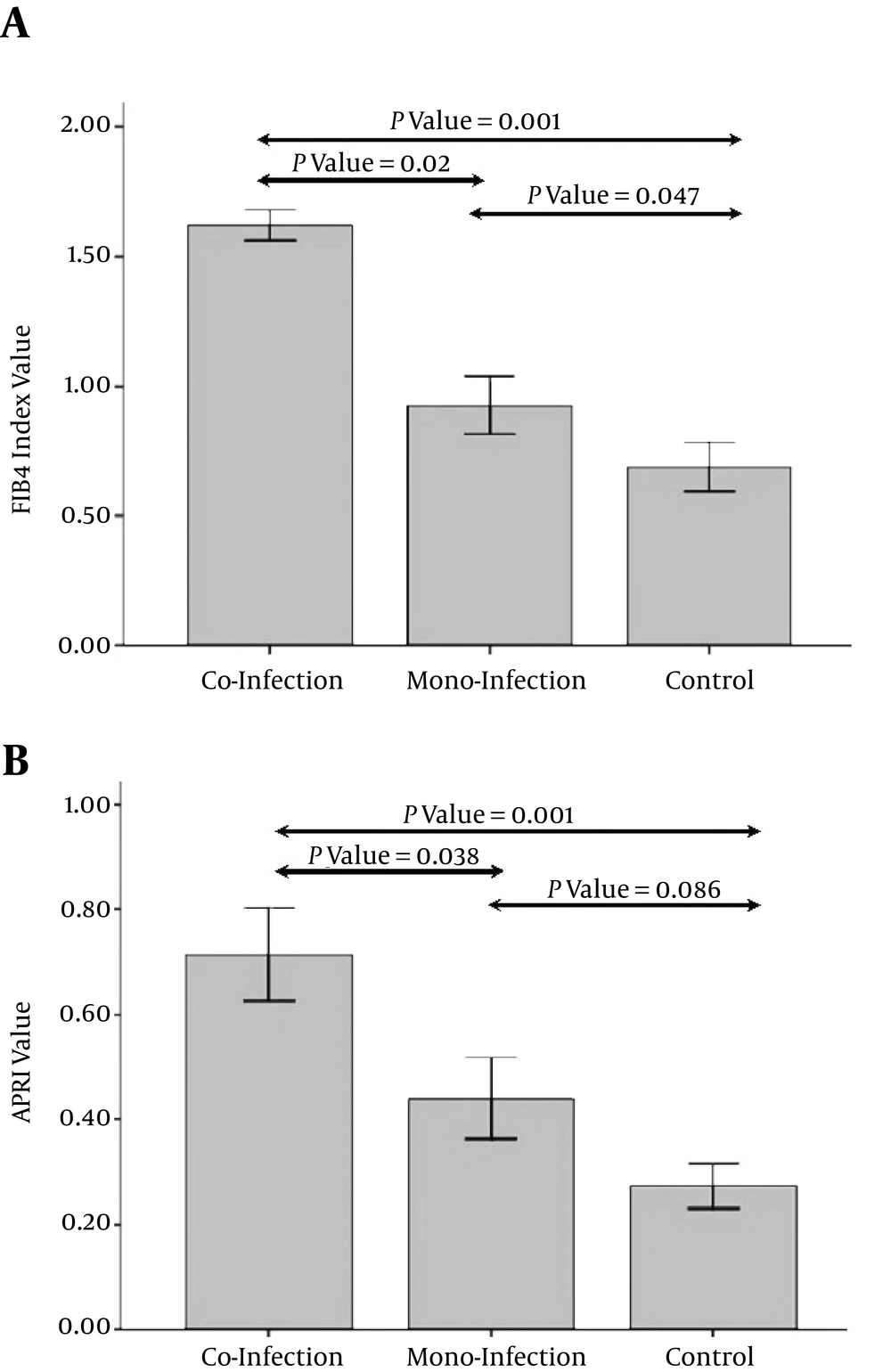
As expected, there were significantly lower CD4+ cell counts in both HIV mono-infection and HIV/HCV co-infection groups compared to the control group (P < 0.05). In addition, although there was a slight decrease in CD4+ cell counts in HIV/HCV co-infected patients, this difference was not statistically significant. Compared to controls, whole blood miR-29a-5p was markedly downregulated in HIV mono-infected patients by 0.679 fold (S.E. range 0.524 - 0.833) and also showed a remarkable decrease in co infected subjects by 0.286 fold (S.E. range 0.198 - 0.312) (Figure 1). On the other hand, a dissimilar trend is observed for the FIB-4 index so that this index is the highest in HIV/HCV co infected patients and the lowest in controls (Figure 2A). Furthermore, a similar alteration is observed with regard to the APRI index in Table 2, where there was a significant increase in the APRI index in HIV/HCV co infected and HIV mono-infected patients compared to healthy controls (see also Figure 2B).
MiR-29 levels (in relative units) are indicated as a block value of 100% for the control group; the percentage decreased to 68% (0.679 fold reduction) for HIV-mono-infected and 28.6% (0,286 fold reduction) for HIV-HCV-co-infected patients which showed a significant difference with P value of P = 0.037.
A, FIB-4 value in studied subjects: the error bar plot started with the control group (FIB-4 index value about 0.7), increased over 0.9 (nearly similar value) for mono-infected and up to 1.6 (2.4 fold increase) for co-infected patients with an acceptable significance (P = 0.001). According to the error plot, there were statistically significant differences between co-infection group and mono-infection and healthy control groups as well as between mono-infection group and healthy control group in terms of mean FIB4 (P < 0.05); B, APRI score in studied subjects: similarly, error bar plot started with the control group (APRI value about 0.27) which increased over 0.44 (1.6 fold increase) in mono-infection group and reached nearly below 0.71 (about 2.6 fold increase) in con-infection group with still an acceptable significance (P = 0.001). According to the error plot, there were statistically significant differences between co-infection group and mono-infection and healthy control groups in terms of APRI index (P < 0.05), whereas there was not any significant difference between mono-infected patients and healthy controls.
As expected, CD4+ cell counts were significantly (P < 0.001) higher in patients under treatment (539.87 ± 283.79 cell/µL) than patients who had not taken any drug (341.98 ± 242.55 cell/µL). Correlation analysis also showed that CD4+ cell counts were directly correlated with consumption of drugs (rs = 0.341, P < 0.001). In addition, there was an ascending trend in miR-29a-5p levels among patients taking medicine (0.99 ± 0.27 fold) compared to the others (0.9 ± 0.36 fold), though this increase was not statistically insignificant (P < 0.395).
Next, we investigated the possible correlation of blood levels of miR-29a-5p with clinical outcomes and laboratory findings in studied groups. Our results showed that decreased levels of miR-29a-5p were accompanied by an increase in aminotransferases. In addition, as shown in Table 3, miR-29a-5p was indirectly correlated with ALT activity indicating a decrease in the level of miR-29a-5p with increasing ALT activities. A similar noticeable reverse correlation was also observed between blood miR-29a-5p levels and APRI index (Table 3). The direct correlation between miR-29a-5p down-regulation and CD4+ cell count was also significant. Furthermore, there was a significant reverse correlation between FIB-4 index and miR-29a-5p level (Table 3).
| APRI | FIB4 | CD4+ | ALT | AST | Hb | ||
|---|---|---|---|---|---|---|---|
| miR-29 | P value | < 0.001 | 0.005 | 0.045 | 0.032 | 0.114 | 0.081 |
| rs | -0.712 | -0.691 | 0.225 | -0.237 | -0.176 | 0.468 |
Abbreviations: rs, spearman correlation coefficient; AST, aspartate aminotransferase; ALT, alanine aminotransferase; Hb, hemoglobin.
5. Discussion
Human immunodeficiency virus greatly decreases immune system responses and also may affect pathological outcome of hepatitis C virus and facilitate its exit from latency status (28-30). Co-infection of HIV with HBV and/or HCV increases the risk of hepatic damages and hepatotoxicity from ART regimen (4). These pathologic changes in co-infected patients are attributed to different molecular mechanisms in these patients compared to mono-infected subjects. In this regard, a different expression of micro RNA in co-infected patients compared to healthy and mono-infected patients has recently been described (31).
In the present study, we showed that miR-29a-5p had statistically significant lower expression in co-infected than mono-infected patients. Besides, our results showed that the expression of miR-29a-5p decreased in both mono- and co-infected patients compared to healthy subjects. Ahluwalia et al. (32) showed that the cellular miR-29 decreases the production of negative regulatory factor protein and inhibits HIV-1 proliferation. In another study, Nathans et al. (13) confirmed that the HIV-1 3′UTR binds to miR-29 as a potential target and inhibits miR-29 and increases HIV-1 reproduction. In a recent study, Patel et al. (33) proved that the miR-29 expression inversely linked to HIV-1 replication in in vitro and in vivo models. They suggested that miR-29 is associated with HIV latency period as a novel approach to eliminate latent HIV-1 pools. It has been recently shown that IL-21 induces the expression of miR-29 in CD4+ cells through STAT3 and that the IL-21/miR-29 axis efficiently suppresses HIV-1 replication. Interestingly, exogenous IL-21 limits early HIV-1 infection and lower viremia in vivo by increasing miR-29 expression (34).
Recently, Kaleji et al. (15) demonstrated a lower expression of miR-29a in HIV patients under HAART and patients resistance to HAART compared to controls. Their study also revealed miR-29b was down-regulated in HIV patients including people without drug treatment, patients under treatment, and drug-resistant patients compared to healthy subjects. They showed that down-regulation of miR-29a and miR-29b is associated with higher viral load and lower CD4+ cell counts. More recently, Rosca et al. (35) showed that miR-29a expression has a reverse correlation with HIV viral load and a direct correlation with CD4+ cell count and CD4+/CD8+ ratio. They showed that patients with failed treatment including those with detectable viremia and those with CD4+ cell count below 350 cells/mm3 have a significant decrease in miR-29a expression. However, they did not find any significant correlation between miR-29a levels and nadir CD4 count or zenith HIV viral load.
In line with the previous studies, our study showed that miR-29 significantly decreased in HIV patients compared to healthy subjects. On the other hand, it seems that a lower expression of miR-29 was correlated with higher HCV load. Bandyopadhyay et al. (14) demonstrated that HCV infection reduces the expression of all members of miR-29 family including miR-29a, miR-29b, and miR-29c in hepatocytes. Furthermore, they showed that down-regulation of miR-29 increases the collagen synthesis in activated liver stellate cells. They suggested that treatment with miR-29 agomirs might decrease liver fibrosis in HCV infection. Importantly, we showed that miR-29a-5p further reduces in co-infected compared to mono-infected patients. The alteration of miRs expression in HIV co-infection with viral hepatitis has been recently studied. Gupta et al. (36) analyzed 847 miRs to determine differentially expressed miRs in peripheral blood mononuclear cells in HIV/HCV co-infected, HIV mono-infected, and HCV mono-infected patients compared to controls. They showed that miR-122 had the highest expression level in HIV/HCV co-infected patients compared to controls. They also revealed that miR-29b and miR-29c were up-regulated in HIV/HCV co-infected patients compared to controls. It has been clearly shown that the level of serum miR-122, miR-22, and miR-34a was correlated with the progression of liver damages in HIV infected patients. It seems that these micro RNAs are well illustrate different ways of liver injury and are independent biomarkers for predicting hepatic damage in HIV patients (31).
We also evaluated the possible association between miR-29 and clinical and laboratory outcomes of the studied groups. In line with earlier studies (20, 23-25), we showed a significant decline in CD4+ cell counts among patients co-infected with HIV and HCV compared to patients with HIV mono-infection and healthy individuals. Additionally, we showed that miR-29 was down-regulated with decreasing CD4+ cell counts in all subjects. An inverse correlation was also observed between miR-29 expression and liver injury. These results are supported by previous findings by Anadol et al. (31), revealing that circulating miRs including miR-122, miR-22, and miR-34a has a correlation with liver injury and fibrosis.
Our study had a limitation. Since the FIB-4 and APRI scores have surely a minor sensitivity and accuracy for liver fibrosis assessment compared to modern fibro scan modalities, future studies are suggested by using this new technique. In summary, we showed a significantly lower expression of total miR-29 in HIV/HCV co-infected patients compared to HIV positive subjects and controls. An inverse correlation of CD4+ cell counts with miR-29 expression was also observed in this study indicating a decrease in miR expression by the reduction of CD4+ cell counts and liver injury in all patients, including HIV positive and HIV/HCV co-infected subjects. It can be concluded that the determination of miR-29 expression might be a distinct biomarker for liver injury in HIV-infected patients.