1. Background
It is a well-established fact that hepatitis C virus (HCV) is playing havoc worldwide. HCV genotypes are substantially divergent in sequence from each other; they fall into seven phylogenetic clades, designated as genotypes and within these, a variable number of sub-groupings are obvious. HCV variants circulating in Western countries have been designated as subtypes of which, 1a, 1b, 2a, 2b, 3a, 4a, and 6a are the most frequently identified ones (1). HCV subtypes are epidemiologically divergent, with differences in risk group targeting and geographical distributions that reflect their recent epidemic spread (2). The rate of nucleotide sequence change in viruses, particularly in those with RNA genome e.g. HCV, HGV/GBV-C, is invariably much greater than that of their hosts; this presents a number of problems in evolutionary reconstruction. This introduces a significant genome diversity in HCV, which necessitates the classification of different strains into major genotypes and their subtypes. Understanding the origin and nature of HCV genetic diversity is critical for defining preventive strategies as well as for development of therapies and vaccine. Phylogenetic methods are essential tools for the study of molecular epidemiology. The confirmatory gold standard and the most definitive method for HCV genotyping is sequencing of a specific PCR-amplified portion of the HCV genome obtained from the patient, followed by phylogenetic analysis (3, 4).
Moreover, beside HCV genotypes, hematological, biochemical, and pre-treatment viral load has also been shown to be the prognostic indicator of response to antiviral therapy (5), as increased pre-treatment viral load and very disturbed hemato-biochemical values have been linked with low rates of response to standard interferon therapy (6). In Pakistan, although considerable research has been observed in many districts and cities on the prevalence of HCV genotypes (7, 8), not much data is available regarding the correlation between hemato-biochemical values, viral load, and HCV genotypes. Therefore, the present study emphasizes the distribution of different HCV genotypes, pretreatment hemato-biochemical analysis, and HCV RNA viral load in HCV infected patients in Rahim Yar khan, Okara, and Toba Tek Singh districts of Punjab province, Pakistan, which are most neglected areas of this region.
2. Methods
2.1. Sample Collection, Serological and Hemato-Biochemical Analysis
A total 1115 blood samples were collected from the three districts of Rahim Yar Khan (n = 439), Okara (n = 317), and Toba Tek Singh (n = 359) in Punjab from the clinically affected patients for one year from 2012 to 2013. According to the inclusion criteria, the study involved only those patients with the clinical signs and results based on ELISA. All the patients with any other hepatitis due to reasons other than hepatitis C (e.g. autoimmune hepatitis, hemochromatosis, and Wilson’s disease) were excluded. Initially, consent was taken from each individual and the study was approved by the ethics committee of the institute. Then, all samples were screened by ELISA followed by Hemato-biochemical analysis (hemoglobin, platelets, TLC, total bilirubin, ALT, ALP, Total proteins, albumin, and globulin) on Hitachi 902 fully automated chemistry analyzer by Roche Diagnostics, Germany.
2.2. HCV Qualitative and Quantitative Test
HCV qualitative and quantitative analysis was carried out for all the samples by the method of Ali et al. 2008 (8). All qualitative PCR positive samples were further subjected for HCV genotyping by the protocol carried by Afridi et al. 2014 (9). Serum from each blood sample was separated at 3000 g, tagged, and kept at -20°C until further processing. RNA extraction was carried out using Qiagen kit (Invitrogen, Corp., California; USA) as per manufacturer’s instruction. Complimentary DNA (cDNA) was synthesized using Invitrogen kit as per manufacturer’s guideline. The final PCR product gained either after qualitative or genotype-specific PCR was further processed to electrophoresis in 2% agarose gel containing ethidium bromide along with 100-bp DNA ladder (Invitrogen, Corp., California; USA). The photographs of banding pattern, was taken by Gel Doc System to check genotype specific bands.
2.3. Statistical Analysis
Statistical analysis was done by using SPSS version 16.0 software. Student t-test and Chi-square tests were performed to analyze differences in continuous and categorical data, respectively. P value < 0.05 was considered as significant. Multiple regression analysis was used to evaluate independent associations between HCV genotypes, individual demographic characteristics, and biochemical values to identify variables association within different genotypes.
3. Results
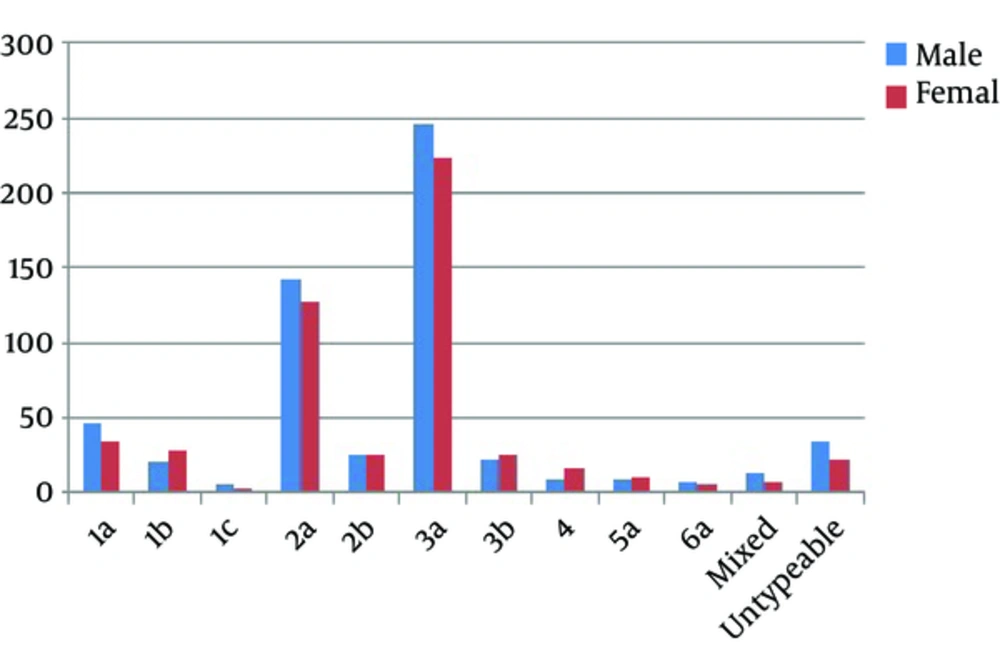
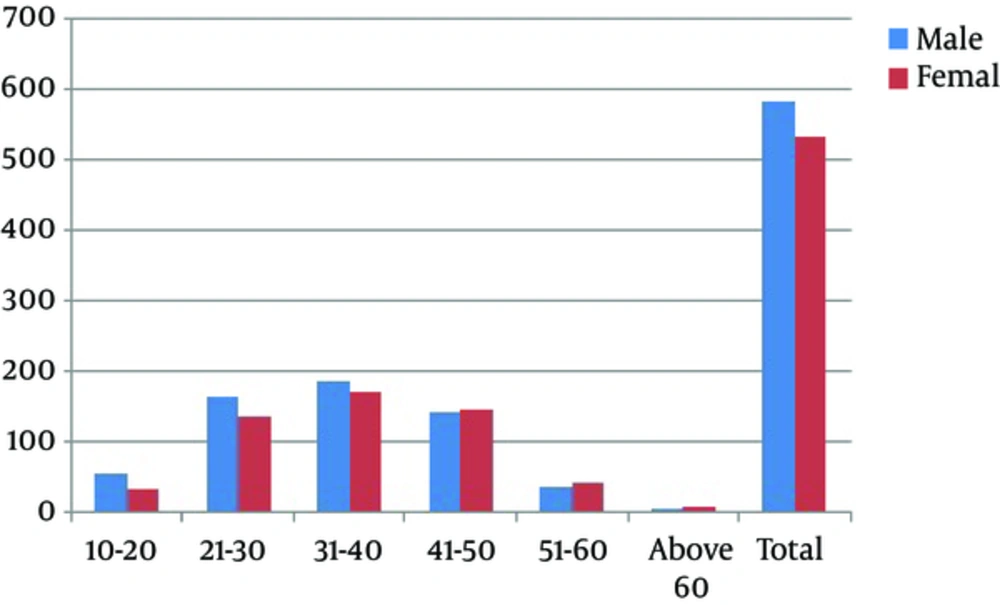
The prevalence of different HCV genotypes and viral load among both genders in different geographical regions of Punjab province is shown in Figure 1 and Table 1. Of 1115 samples, 583 (52.3%) from males and 532 (47.7%) from females were declared as HCV positive that showed nearly equal representation in both genders (Figure 2). Among these 1115 samples, 1057 (94.8%) were detected based on specific PCR fragment while 58 (5.2%) remained unable to demonstrate a specific genotypic band. Genotype 3 (46.3%) was the most frequent one in both genders followed by genotype 2 (28.9%), genotype 1 (12.5%), genotype 4 (2.2%), genotype 5 (1.8%), genotype 6 (1.2%), and untypeable genotypes (5.2%) (Figure 1). Among the sub-genotypes, the most detected sub-genotype in males was 3a subtype (42.20%) followed by 2a subtype (24.53%) and 1a subtype (7.89%). Other less frequent subtypes were 2b (2.49%), 3b (3.77%), 1b (3.6%), 1c (1.03%), 4 (1.54%), 5a (1.54%), 6a (1.20%), and mixed (2.4%). In females, the most frequent sub-genotypes were 3a (41.92%), 2a (24.06%), 1b (5.26%), 3b (4.70%), 1c (0.56%), 4 (3.01%), 6a (1.13%), mixed (1.4%), and 2b (4.89%) (Figure 1).
| Genotype/Subtype | Viral Load | P Value | ||
|---|---|---|---|---|
| < 600,000 | 600,000 - 800,000 | > 800,000 | ||
| Genotype 3 | 170 | 196 | 150 | 0.937 |
| Other genotypes | 194 | 225 | 180 | |
| Male | 186 | 239 | 158 | 0.065 |
| Female | 188 | 182 | 162 | |
| Toba Tek Singh | 0.301 | |||
| Genotype 3 | 53 | 47 | 51 | |
| Other genotypes | 74 | 78 | 56 | |
| Okara | 0.454 | |||
| Genotype 3 | 37 | 63 | 37 | |
| Other | 60 | 73 | 47 | |
| Rahim Yar Khan | 0.475 | |||
| Genotype 3 | 59 | 72 | 50 | |
| Other | 91 | 88 | 79 | |
Based on age, different groups of genotyped samples were formed such as 10 - 20 years, 21 - 30 years, 31 - 40 years, 41 - 50 years, 51 - 60 years, and above 60 years. The overall percentage of positive HCV samples in all the age groups was higher in males than females except for the age group 41 - 50 years, 51 - 60 years, and above 60 years (Figure 2). The distribution of genotypes in all the age group categories was also observed. Genotype 3a was highest in all the age groups (42.1%) followed by genotypes 2a (24.3%), 1a (7.3%), 2b (4.6%), 1b (4.4%), 3b (4.2%), 4 (2.2%), 5a (1.8%), 6a (1.2%), and mixed genotypes (2.0%). On the other hand, the age group 31 - 40 years had the highest frequency of cases while the lowest frequency was observed in the age group of above 60 years (Table 2).
| Genotype | Age Group | |||||
|---|---|---|---|---|---|---|
| 10 - 20 | 21 - 30 | 31 - 40 | 41 - 50 | 51 - 60 | Above 60 | |
| 1a | 4 (4.60) | 21 (7.00) | 23 (6.46) | 25 (8.77) | 8 (10.26) | 0 (0) |
| 1b | 5 (5.75) | 9 (3.00) | 20 (5.62) | 13 (4.56) | 2 (2.56) | 0 (0) |
| 1c | 2 (2.30) | 3 (1.00) | 4 (1.12) | 0 (0) | 0 (0) | 0 (0) |
| 2a | 19 (21.84) | 71 (23.67) | 89 (25) | 66 (23.16) | 23 (29.49) | 3 (33.33) |
| 2b | 5 (5.75) | 10 (3.33) | 20 (5.62) | 13 (4.56) | 2 (2.56) | 1 (11.11) |
| 3a | 39 (44.83) | 142 (47.33) | 136 (38.20) | 116 (40.70) | 33 (42.31) | 3 (33.33) |
| 3b | 6 (6.90) | 14 (4.67) | 15 (4.21) | 7(2.46) | 5 (6.41) | 0 (0) |
| 4 | 2 (2.30) | 5 (1.67) | 7 (1.97) | 10(3.51) | 0 (0) | 1 (11.11) |
| 5a | 2 (2.30) | 2 (0.67) | 9 (2.53) | 6 (2.11) | 1 (1.28) | 0 (0) |
| 6a | 0 (0) | 3 (1) | 7 (1.97) | 2 (0.70) | 1 (1.28) | 0 (0) |
| Mixed | 2 (2.29) | 6 (2) | 6 (1.68) | 7 (2.45) | 1 (1.28) | 0 (0) |
| Untypeable | 1 (1.15) | 14 (4.67) | 20 (5.62) | 20 (7.02) | 2 (2.56) | 1 (11.11) |
| Total | 87 | 300 | 356 | 285 | 78 | 9 |
aValues are expressed as No. (%).
Among the total HCV positive samples, the highest frequency of HCV positive patients was observed in Rahim Yar Khan with 439 (39.4%) positive cases, followed by 359 (32.2%) that belonged to Toba Tek Singh while the lowest frequency was observed in Okara district with 317 (28.4%) positive samples. Among all the districts, 3a had the highest frequency and 1c had the lowest frequency (Table 3).
| Genotype | Districts | ||
|---|---|---|---|
| Toba Tek Singh | Rahim Yar Khan | Okara | |
| 1a | 27 (7.52 ) | 34 (7.74) | 20 (6.31) |
| 1b | 14 (3.90) | 22 (5.01) | 13 (4.10) |
| 1c | 4 (1.11) | 2 (0.46) | 3 (0.95) |
| 2a | 89 (24.79) | 111 (25.28) | 71 (22.40) |
| 2b | 13 (3.62) | 19 (4.33) | 19 (5.99) |
| 3a | 151 (42.06) | 181 (41.23) | 137 (43.22) |
| 3b | 19 (5.29) | 15 (3.42) | 13 (4.10) |
| 4 | 12 (3.34) | 8 (1.82) | 5 (1.58) |
| 5a | 5 (1.39) | 8 (1.82) | 7 (2.21) |
| 6a | 4 (1.11) | 5 (1.14) | 4 (1.26) |
| Mixed | 8 (2.23) | 8 (1.82) | 6 (1.89) |
| Untypeable | 13 (3.62) | 26 (5.92) | 19 (5.99) |
| Total | 359 (32.30) | 439 (39.37) | 317 (28.43) |
aValues are expressed as No. (%).
Viral load was categorized into three classes according to the specified levels as low level (< 60, 0000 IU/ml), intermediate level (60,0000 - 80,0000 IU/mL), and high level (> 80,0000 IU/mL) viral loads. No significant differences were detected in HCV infected patients of both genders (Table 1). The viral load with respect to genotypes based on districts and genders was determined as shown in Table 1.
Along with the viral load, clinical profile of the patients was also recorded. The clinical profile included different blood parameters, levels of different liver enzymes, and levels of albumin and globulin as given in Table 4.
| Clinical Parameters | Normal Values | Minimum | Maximum | Mean ± Std. Deviation |
|---|---|---|---|---|
| Haemoglobin Level, g/dL | 11 - 18 | 8.10 | 17.80 | 13.59 ± 1.61 |
| TLC Level, * 103/mm3 | 4 - 11 | 2.70 | 27.40 | 9.71 ± 3.48 |
| Platelet Count, *103/mm3 | 150 - 400 | 7.70 | 389.00 | 274.02 ± 51.80 |
| Total Bilirubin, mg/dL | 1 | 0.30 | 4.40 | 1.31 ± 0.65 |
| ALT | < 45 | 21.00 | 978.00 | 71.46 ± 98.02 |
| ALP | < 258 | 106.00 | 735.00 | 242.11 ± 72.27 |
| Total Protein, g/dL | 6.6 - 8.8 | 4.20 | 8.70 | 7.58 ± 0.72 |
| Albumin, g/dL | 3.8 - 5.1 | 1.90 | 5.20 | 4.39 ± 0.63 |
| Globulin, g/dL | 2.3 - 3.8 | 2.10 | 3.90 | 3.19 ± 0.27 |
| Albumin Globulin Ratio | 1.2 - 2.2 | 0.60 | 2.30 | 1.37 ±0.22 |
| ELISA | 1 | 0.29 | 65.40 | 6.73 ± 5.51 |
4. Discussion
The current study demonstrated the frequency of various HCV genotypes in different categories and their association with demographic and biochemical parameters. This study ascertained that HCV genotypes were spread with equal probability between the both sexes and there was no association between gender and HCV genotypes. These results are in agreement with multiple previous studies indicating little statistical association between genders and genotypes in Pakistan (8, 10-12). This study contradicts a Libyan study that showed the association of various HCV genotypes with gender (13).
Regarding age groups, a high prevalence of HCV infection was found to occur below the age of 50 years. The former studies noted a higher prevalence in age group of ≤ 40 years that is in partial agreement with the current findings. It has also been found that HCV infection was higher among females with the age above 40 years as compared to males with the same age.
It has been well-established that HCV genotype 3a is highly prevalent in Pakistani population (9, 11, 14). On the other hand, a single study revealed that 1a genotype has a higher frequency in Balochistan (7). This study inferred that HCV genotype 3a is the most frequent genotype in Pakistan (7, 11). The distribution of HCV genotypes is close to the documented studies from other South Asian countries where genotype 3 is more frequent than other genotypes (15). This study also reported many rare genotypes like 5a, 1c, and 6a, which are uncommon and less reported. Unlike Ali et al. (8) sudy findings, the findings of this study support the verdict of some previous studies that showed the presence of genotypes such as 5a, 4a, and 1c in spite of their rare status (9-11, 16). These findings are in contradiction with those of some studies that have shown the partial absence of genotypes 4, 5, and 6 (8, 17).
Moreover, this study also documented the mixed genotype infections. To the author’s knowledge, mixed genotypes like 1b + 4, 2a + 5a, and 3a + 5a were not reported before in Pakistan. Although the frequency of mixed genotypes was very low in this study, such pattern of co-infections can pose problems in therapeutic regimes (18).
In this study, HCV RNA titre was also determined with respect to genotypes in different districts and genders as shown in Table 1. As determined in the past studies, HCV RNA titer was categorized into three levels based on their specified values as low level (< 600,000 IU/mL), intermediate level (600,000 - 800,000 IU/mL), and high level (> 800,000 IU/mL) (8). Genotype 3 HCV-infected patients falling in the low level RNA titer category i.e. low level (< 600,000 IU/mL) usually get shorter therapy and quickly attain early viral response (EVR) and Sustained Viral Response (SVR) as compared to other categories specially the category of high level (> 800,000 IU/mL) (9, 19, 20). In the current study, about 150 patients from genotype 3 belonged to the low-level RNA titer category, likely to gain undetectable viral load in a 4 to 8-week duration. For the other two categories, the protocols of 16 weeks and 24 weeks are likely to be followed (8, 19, 20). In line with previous studies, this study also emphasizes the need of prior information regarding RNA titer while formulating the therapy protocols including protocols of 16 weeks and 24 weeks (8, 9, 21).
In contrast to most previous studies conducted in Pakistan, this study also reported the clinical profile of HCV patients (22). This study could be used as an evident document in providing information about clinical profile of HCV patients. Moreover, the current study has also tried to correlate the enzyme profile with genotypes recorded. In the former studies, genotypes have not found to be correlated with enzyme profile of the patients (4, 23, 24). Possibly, due to less sample size, regression model and correlation could not provide any statistical significance, which is in contrast to recent studies (16).