1. Background
Hepatitis E virus (HEV) is a major cause of acute viral hepatitis in developing countries (1, 2).
This non-enveloped virus belongs to the Hepevirus genus of the Hepeviridae family. The viral genome is represented by a positive-sense single-stranded RNA of about 7.2 kb that contains three partially overlapping open reading frames (ORFs). ORF1 encodes a non-structural protein with different enzymatic activities (RNA-dependent RNA polymerase, RNA helicase, and protease), ORF-2 encodes the capsid protein, and ORF-3 (which overlaps ORF-2) encodes a viral protein used in virion morphogenesis and release (3, 4). HEV has been classified into four main genotypes [1 - 4] and a number of subtypes [5], although other new genotypes have recently been identified in various mammals (5-7).
The geographical and host-range distribution of HEV genotypes differ. Genotypes 1 and 2 are restricted to humans and are typically fecal-orally transmitted. Genotype 1 is the main cause of sporadic and epidemic hepatitis E in developing regions of Asia, Africa, and South America, while genotype 2 has thus far been identified in patients in Mexico, Chad, and Nigeria (8-10). Genotypes 3 and 4 have been recovered from humans, pigs, and other species and are responsible for sporadic cases of HEV in humans (11-14). HEV genotype 3 has a worldwide distribution and is widespread among pigs in developed countries (3). Evidence now indicates that HEV-3 infection can be transmitted through the ingestion of raw or undercooked meat from infected animals, thereby highlighting the zoonotic nature of this infection (15). Genotype 4 is mainly found in eastern Asia (16).
In Italy, HEV has been found in pigs, boars, and humans (11-13, 17, 18). The relatively high seroprevalence of HEV in domestic pigs indicates an active circulation of HEV in Italy (19). Genotype 3 is the only genotype that has been reported in Italian pigs and in wild boars (20, 21). Caruso et al. who performed a serological and virological survey of HEV in wild boar populations in northwestern Italy, reported a seroprevalence of 4.9% and detected HEV RNA in 3.7% of liver samples, while no serum samples were positive for HEV RNA. Phylogenetic analysis of the ORF2 region revealed that the isolates were clustered within genotype 3, subtypes 3e and 3f, and were closely related to HEV strains previously described in domestic pigs from the same geographic area (14).
Genotyping of the HEV virus in association with the evolutionary rate estimate by phylogenetic analysis can aid in determining the circulation of the virus and in understanding viral evolution and the mechanism of infection.
2. Objectives
The aim of this study was to investigate the genetic diversity of HEV in Italy in order to obtain a more in-depth insight into the phylogenetic relationships among different strains of genotype 3, the most frequent circulating in Italy, among humans and swine. An additional aim was to estimate the date of origin and the demographic history of HEV circulation in Italy.
3. Materials and Methods
3.1. Data Set
A total of 327 HEV sequences of swine and humans from Italy were downloaded from the national centre for biotechnology information (http://www.ncbi.nlm.nih.gov/). Three different data sets were built: the first contained 11 sequences of HEV from Italy from the human ORF2 capsid gene, genotype 3, plus 13 genotype-specific reference sequences. The second data set contained 65 sequences of HEV from Italy from the swine ORF2 capsid gene, genotype 3, plus 13 genotype specific reference sequences. The third data set contained the 76 ORF2 capsid gene sequences of the HEV genotype 3 from humans and swine from Italy.
The first and the second data sets were used to confirm the genotype of the analyzed sequences. The first and the second datasets were analyzed separately, without reference sequences, to conduct a selective pressure analysis and to obtain demographic history in both humans and swine. The third data set was used to estimate the mean evolutionary rate, to perform the time-scaled phylogeny, and to obtain the demographic history.
All the reference sequences were downloaded from the national centre for biotechnology information (http://www.ncbi.nlm.nih.gov/). The reference sequences were selected based on the following inclusion criteria: 1) sequences already published in peer-reviewed journals; 2) no uncertainty about genotype/subtype assignment; 3) sampling dates were known and clearly established in the original publication.
3.2. Likelihood Mapping
The phylogenetic signal in a data set of aligned DNA or amino acid sequences can be investigated with the likelihood mapping method by analyzing groups of four randomly chosen sequences, called quartets (22). A quartet has three possible unrooted tree topologies. The likelihood of each topology is estimated with the maximum likelihood method and the three likelihoods are reported as a dot in an equilateral triangle (the likelihood map).
Three main areas can be distinguished in the map: the three corners representing fully resolved tree topologies (i.e., the presence of a treelike phylogenetic signal in the data), the center (which represents a star-like phylogeny), and the three areas on the sides that indicate a network-like phylogeny, (i.e., the presence of recombination or conflicting phylogenetic signals). A substantial star-like signal (i.e., a star-like outburst of multiple phylogenetic lineages) is indicated by > 33% dots falling within the central area, as confirmed by extensive simulation studies. Likelihood mapping analyses have been performed with the TREE-PUZZLE program by analyzing 10,000 random quartets (22).
3.3. Phylogenetic Analysis
The sequences of all datasets were aligned using Clustal X and manually edited by Bioedit (23). The genotype of the Italian sequences was determined by phylogenetic analysis. The maximum likelihood (ML) phylogenetic tree was generated with the HKY + I + G model of nucleotide substitution, using Phyml v 3.0 (23, 24). The evolutionary model was chosen as the best-fitting nucleotide substitution model, based on the results of the hierarchical likelihood ratio test (HLRT) implemented in Model-test software version 3.7 (25). The statistical robustness and reliability of the branching order within the phylogenetic trees was confirmed by bootstrap analysis, considering a bootstrap value > 70% as significant statistical support.
3.4. Bayesian Phylogenetic Analysis: Evolutionary Rate Estimate, Dated Tree, and Demographic History
The evolutionary rate and the dated tree for the third data set was co-estimated using a Bayesian Monte Carlo Markov Chain (MCMC) approach that implemented the HKY + I + G model and used both a strict and an uncorrelated log-normal relaxed clock model. Three parametric demographic models of population growth (constant size, exponential, and expansion) and a Bayesian skyline plot (BSP, a non-parametric piecewise-constant model) were compared as coalescent priors. The best fitting models were selected by means of a Bayes factor (BF, using marginal likelihoods) implemented in Beast v. 1.7.4 (23).
In accordance with Villano et al. (26), the strength of the evidence against H0 (null hypothesis) was evaluated as follows: 2lnBF < 2 = no evidence; 2 - 6 = weak evidence; 6 - 10 = strong evidence; and >10 = very strong evidence. A negative 2lnBF indicates evidence in favor of H0. Only values ≥ 6 were considered significant. The MCMC chains were run for at least 50 million generations and were sampled every 5,000 steps.
Convergence was assessed by estimating the effective sampling size (ESS) after a 10% burn-in, using Tracer software, version 1.5 (http://tree.bio.ed.ac.uk/software/tracer/), and accepting ESS values of 250 or more.
Uncertainty in the estimates was indicated by 95% highest posterior density (95% HPD) intervals. Statistical support for specific clades was obtained by calculating the posterior probability of each monophyletic clade.
The obtained tree was summarized by Tree Annotator (included in the Beast package) by choosing the tree with the maximum product of posterior probabilities (maximum clade credibility or MCC) after a 10% burn-in.
The demographic history was analyzed on the first and second dataset by performing the Bayesian skyline plot.
3.5. Selective Pressure Analysis
The CODEML program implemented in the PAML 3.14 software package (http://abacus.gene.ucl.ac.uk/software/ paml.html) was used to investigate the adaptive evolution of the HEV capsid gene. The sequences alignments of the first and second dataset were used to test whether they were under positive selection.
The following six models of codon substitution were used in this analysis (27): M0 (one-ratio), M1a (nearly neutral), M2a (positive selection), M3 (discrete), M7 (beta), and M8 (beta and omega). These models are nested, so we used codon-substitution models to fit the model to the data, using the likelihood ratio test (LRT) (28). The discrete model (M3), with three dn/ds (ω) classes, allows ω to vary among sites by defining a set number of discrete site categories, each with its own ω value. Maximum-likelihood optimization allows the estimation of the ω and P values and the fraction of sites in the aligned data set that falls into a given category. Finally, the algorithm calculates the a posteriori probability of each codon belonging to a particular site category. The M3 model was then used to designate sites with a posterior probability exceeding 90% and a ω value > 1.0 as being “positive selection sites” (29). The site rate variation was evaluated by comparing M0 with M3, while positive selection was evaluated by comparing M1 with M2. The Bayes empirical bayes (BEB) approach, implemented in M2a and M8, was used instead to determine the positively selected sites by calculating the posterior probabilities (P) of ω classes for each site (30). Note that PAML LRTs have been reported to be conservative for short sequences (e.g., positive selection could be underestimated), although the Bayesian prediction of sites under positive selection is largely unaffected by sequence length (28, 29).
The dN/dS rate (ω) was also estimated by the ML approach implemented in the program HyPhy (31). In particular, the global (assuming a single selective pressure for all branches) and the local (allowing the selective pressure to change along every branch) models were compared by the likelihood ratio test (LRT). Site-specific positive and negative selection were estimated by two different algorithms: the fixed-effects likelihood (FEL), which fits an ω rate to every site and uses the likelihood ratio to test if dN = dS; and the random effect likelihood (REL), a variant of the Yang-Nielsen approach (27), which assumes that a discrete distribution of rates exists across sites and allows both dS and dN to vary independently, site by site. The three methods have been described in more detail elsewhere (31, 32). Sites were selected under selective pressure and our test was kept conservative by assuming a P value of ≤ 0.1 or a posterior probability of ≥ 0.9 as relaxed critical values (31). Part of the analysis was conducted by using the web-based interface Datamonkey (http://www.datamonkey.org/) (31).
The evolutionary analysis was conducted using the reference sequence with accession number AB369687.1 (complete genome cds) to trace the exact position of the amino acids under selection.
4. Results
4.1. Likelihood Mapping
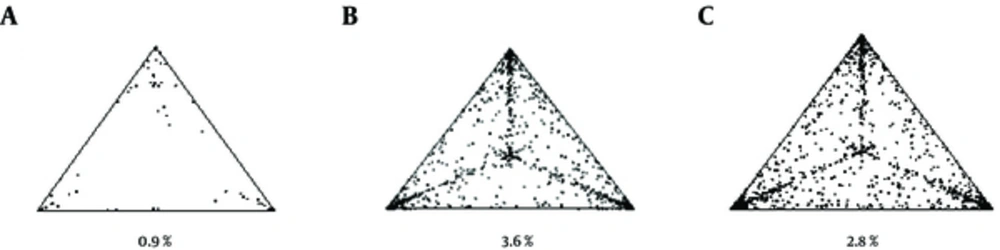
The phylogenetic noise of each data set was investigated by likelihood mapping (Figure 1). The percentages of dots falling in the central area of the triangles were 0.9% (panel a), 3.6% (panel b), and 2.8% (panel c) for the first, second, and third data sets, respectively. None of the datasets showed more than 33% noise, so they contained sufficient phylogenetic signal.
Likelihood mapping of the A, first; B, second and C, third data sets. Each dot represents the likelihood of the three possible unrooted trees for a set of four sequences (quartets) selected randomly from the data set: dots close to the corners or the sides represent tree-like or network-like phylogenetic signals, respectively, in the data. The central area of the likelihood map represents a star-like signal. The percentage of dots in the central area is given at the base of each map.
4.2. Phylogenetic Analysis
Maximum Likelihood phylogenetic trees of the first and second data set showed that all the sequences analyzed in this study were classified as genotype 3 (data not shown). The phylogenetic relationships among the different sequences of HEV were supported by bootstrap analysis, with values > 70%.
4.3. Bayesian Phylogenetic Analysis: Evolutionary Rate Estimate, Dated Tree, and Demographic History
The BF analysis showed that the data were significantly better fitted with the relaxed clock than with the strict clock (2 lnBF = 56.49 for relaxed clock). The BF analysis under the relaxed clock showed that the exponential growth model was better than the other models (2lnBF > 15.736). The estimated mean value of the HEV capsid gene evolutionary rate was 3.9 × 10-3 substitutions/site/year (95% HPD: 1.3 × 10-3 - 7.0 × 10-3).
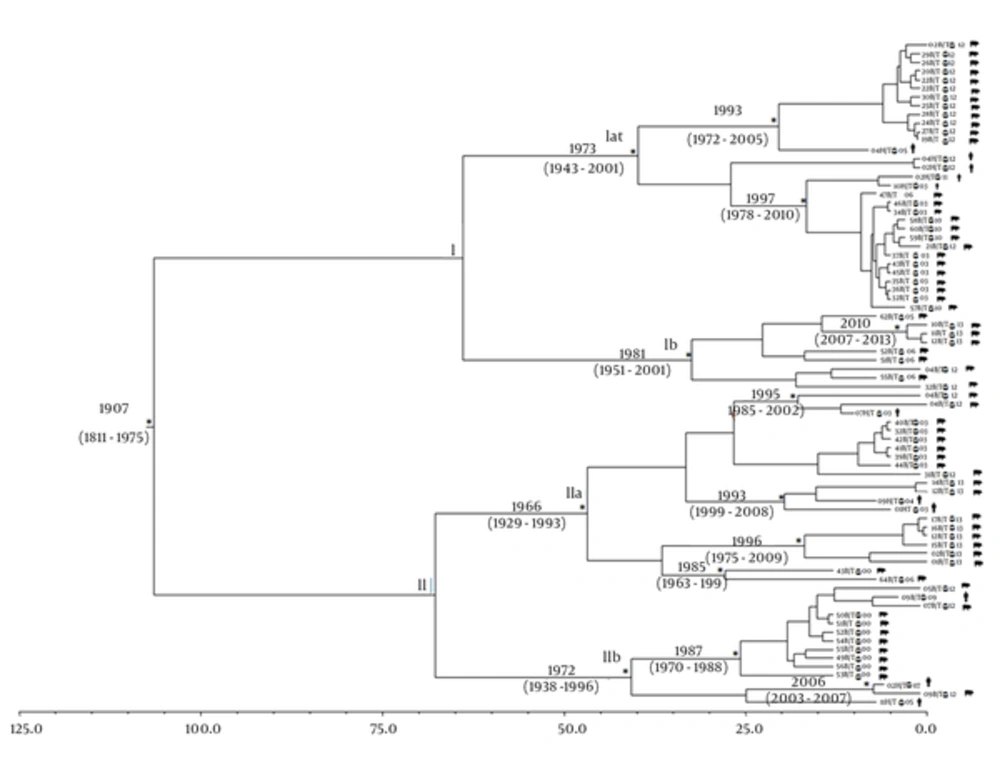
Figure 2 shows the Bayesian maximum clade credibility tree and the time of the most common recent ancestor (tMRCA) estimates performed on the third data set. The root of the tree had a tMRCA of 106 years, corresponding to the year 1907 (95% HPD: 1811 - 1975). Two main clades (clade I and II) were found.
The asterisks (*) along the branches represent significant statistical support for the clades subtending those branches (posterior probability > 0.98). The scale at the bottom of the tree represents time in years. Main clades and clusters are indicated. Human and swine sequences are indicated with different symbols next to the tips of the sequences.
Clade I includes 40 sequences, 35 from swine and 5 from humans, divided into two sub-clades (Ia and Ib). Sub-clade Ia dated back to the year 1973 (95% HPD: 1943 - 2001) and included two other statistically supported clusters: the first dated back to the year 1993 (95% HPD: 1972 - 2005) and the second dated back to the year 1997 (95% HPD: 1978 - 2010) and the sequences from swine and humans appeared closely related. Sub-clade Ib dated back to the year 1981 (95% HPD: 1951 - 2001) and included only one statistically supported cluster, which dated back to the year 2010 (95% HPD: 2007 - 2013) and consisted of only swine sequences. Clade II included 36 sequences (30 from swine and 6 from humans) divided into sub-clades (IIa and IIb). Sub-clade IIa dated back to the year 1966 (95% HPD: 1929 - 1993) and included four statistically supported clusters. The first cluster dated back to the year 1995 (95% HPD: 1985 - 2002), while the second dated back to the year 1993 (95% HPD: 1999 - 2008) and included closely related sequences from swine and humans; the third and the fourth clusters dated back to 1996 (95% HPD: 1975 - 2009) and 1985 (95% HPD: 1963 - 1999), respectively, and included only sequences of swine origin.
Sub-clade IIb dated back to the year 1972 (95% HPD: 1938 - 1996) and included two statistically supported clusters: the first dated back to the year 1987 (95% HPD: 1970 - 1988) and the second to the year 2006 (95% HPD: 2003 - 2007); in these two clusters, only three sequences were from humans.
4.4. Population Dynamics
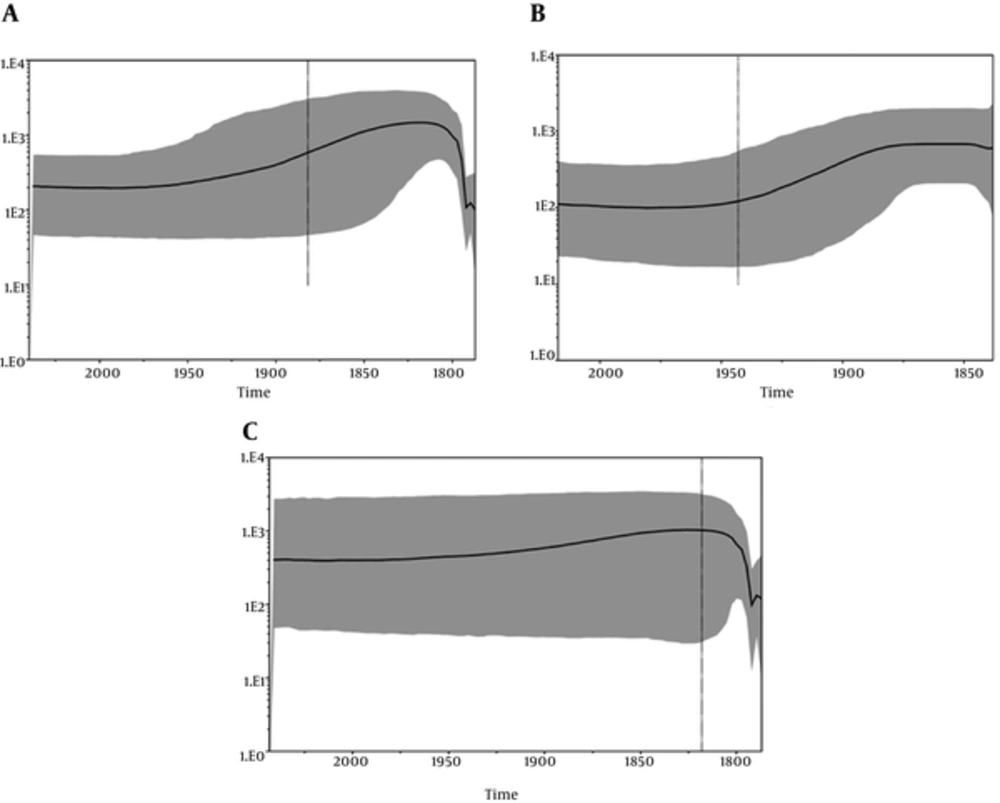
Analysis of the skyline plot (Figure 3A) showed that the effective number (Ne) of HEV infections from human and swine, analyzed together, started to grow approximatively in the early 1900s and reached a plateau in 2000. At the plateau, the epidemic stopped growing but remained at a level higher than it had been at the beginning, even though a decreasing phase, showing a typical “bottleneck,” was evident immediately after 2000.
A, Bayesian skyline plot (BSP) of the HEV Human and swine ORF2 capsid gene sequences from Italy. The effective number of infections is reported on the Y-axis. Time is reported in the X-axis. The colored area corresponds to the credibility interval based on the 95% highest posterior density interval (HPD); B, Bayesian skyline plot (BSP) of the HEV human ORF2 capsid gene sequences from Italy. The effective number of infections is reported on the Y-axis. Time is reported in the X-axis. The colored area corresponds to the credibility interval based on the 95% highest posterior density interval (HPD); C, Bayesian skyline plot (BSP) of the HEV swine ORF2 capsid gene sequences from Italy. The effective number of infections is reported on the Y-axis. Time is reported in the X-axis. The colored area corresponds to the credibility interval based on the 95% highest posterior density interval (HPD).
Skyline plot analysis was also performed separately for human and swine sequences (Figure 3B and C) to determine if the bottleneck evidenced in early 2000 was present in human as well as swine infections. The separated analysis demonstrated the presence of this bottleneck only in the skyline plot from the swine sequences (Figure 3C).
4.5. Evolutionary Analysis
Selection pressure analysis performed on the first data set (sequences isolated from Human) did not reveal any positively selected sites that were statistically supported (using both HYPHY and PAML). The Alfa parameter of the gamma distribution was < 1 and showed a characteristic L-shape, which suggested a nucleotide substitution rate heterogeneity across sites but with most sites highly conserved. Regarding the selective pressure analysis on the first data set, the average ω ratio ranged from 0.0083 to 0.0086 among all models, suggesting that a non-synonymous mutation had around 0.83% - 0.86% as much chance as a synonymous mutation of being fixed in the population. Negative selection analysis identified 87 statistically supported sites using FEL (Table 1).
| Analysis | Site |
|---|---|
| Negatively selected sitesa (w for sites < 1) HYPHY software | 1994 (V); 1996(L,F); 1997(C,F); 1998(I); 2000(G); 2001(S); 2002(P); 2003(N); 2004(S); 2005(Y); 2006(T); 2007(N,Y); 2008(T); 2009(P);2010(Y); 2011(T); 2014(A); 2015(L); 2016(G); 2017(L); 2019(D); 2020(A); 2021(L); 2022(E,G); 2023(L); 2024(E); 2025(R); 2026(N); 2027(L); 2028(P); 2034(T); 2036(T); 2038(V); 2040(R); 2042(T,S); 2043(S); 2044(T); 2045(A); 2046(R); 2047(H); 2048(R); 2049(L); 2050 (R); 2051(R); 2052(G); 2053(A); 2054(D); 2055(G); 2056(T); 2057(A); 2058(E); 2059(L); 2060(T); 2061(T); 2062(T); 2063(A); 2065(T); 2066(R); 2067(F); 2069(K); 2070(D); 2071(L); 2072(H); 2073(F); 2075(G); 2076(T,M); 2077(N); 2078(G); 2079(V); 2080(G); 2082(V); 2085(G); 2086(I); 2087(A); 2088(L); 2089(T,I); 2090(L); 2091(F); 2092(N); 2095D); 2096(T); 2098(L); 2099(G); 2100(G); 2101 (L); 2105(L); 2108(S). |
aNegatively selected sites are numbered according to the amino acid position of the capsid protein of HEV isolate accession number AB369687.1
Similarly, selective pressure analysis performed on the second data set (sequences isolated from swine) showed that the Alfa parameter of the gamma distribution was < 1, indicating that this distribution also had a characteristic L-shape, suggesting a nucleotide substitution rate heterogeneity across sites. The average ω ratio ranged from 0.0339 to 0.0389 among all models, which suggested that a non-synonymous mutation had only around 3.39% - 3.89% as much chance as a synonymous mutation of being fixed in the population. The capsid protein of the second data set also showed negative selection: specifically, 80 statistically supported, negatively selected sites were identified by HYPHY (Table 2).
| Analysis | Site |
|---|---|
| Negatively selected sitesa (w for sites < 1) HYPHY software | 2011(Y,H); 2012(T,P); 2013(G); 2014(A); 2015(L); 2016(G); 2017(L); 2018(L); 2019(D); 2020(F); 2021(A); 2022(L); 2023(E); 2024(L); 2025(E); 2026(F); 2027(R,I); 2028(N); 2029(L,Q); 2030(T); 2031(P); 2032(G); 2033( N); 2034(T); 2035(N); 2036(T); 2037(R);2040(R); 2041(Y); 2042(T); 2043(S); 2044(T); 2045(A,T); 2046(R); 2047(H); 2049(L); 2050(R,G); 2051(R); 2052(G); 2053(A); 2054(D); 2055(G); 2056(T); 2057(A); 2058(E); 2059(L); 2060(T); 2061(T); 2062(T); 2063(A); 2065(T); 2066(R,H); 2067(F); 2070(D;E); 2071(L); 2072(H,L); 2073(F); 2074(T,I); 2075(G,E); 2076(T,M); 2077(N,K); 2078(G); 2079(V,I); 2080(G,R); 2081(E,K); 2082(V,G); 2083(G); 2084(R,L); 2085(G); 2086(I,V,L,R); 2088(L); 2089(T); 2090(L,P,K); 2091(F); 2092(N); 2095(D); 2096(T); 2099(G); 2100(G); 2101(L). |
aNegatively selected sites are numbered according to amino acid position of capsid protein of HEV isolate accession number AB369687.1
5. Discussion
HEV infection is a global cause for morbidity and mortality. Besides endemic infections, autochthonous infections in developed countries are frequent (33). The transmission route is one of the most discussed issues about HEV, and marked differences are observed in different geographical areas. In developed countries, two main transmission routes are described: the fecal-oral route associated with genotypes 1 and 2 transmission and the transmission route through the ingestion of raw meat of infected animals, associated with genotypes 3 and 4 transmission (34).
This study analyzed the phylogenetic relationships among different strains of HEV genotype 3, circulating in Italy between humans and swine, to estimate the date of origin, the spread, and the demographic history of the HEV epidemic in Italy. Our estimate of capsid gene evolutionary rate was 1.8 × 10-3 substitutions/site/year, with a broad credibility interval (between 1.2 × 10-3 and 5.0 × 10-3) that is similar to the value reported by other authors (34). Based on this temporal reconstruction, we suggest that the HEV genotype 3 strains circulating in Italy in the first decade of 1900 diverged into the two main clades I and II, which include the subclades Ia, Ib, and IIa, IIb, respectively, which originated between 1966 and 1981. Interestingly, all swine sequences cluster together, except in some cases where they are intermixed with human sequences, as expected.
Skyline plot analysis was performed globally as well as separately for human and swine sequences. The separate analysis revealed the presence of a bottleneck after the year 2000 but only in the swine sequence data set. This could be due to swine slaughter, which consequently decreased the the number of swine infections registered. These data could suggest that the control of HEV infection depends on adequate measures of prevention to avoid infection of farmers through contact with swine meat and the consequent spread of the virus among humans.
Selective pressure analysis was also used to investigate the presence of sites under negative and positive selection. An average ω ratio < 1 was found in both the human and swine Italian ORF2 capsid gene sequence datasets, and only statistically supported negatively selected sites were identified; this finding confirms the stability of this viral protein. The HEV evolution, until now, has been characterized by neutral genetic drift. More studies are needed to examine zoonotic transmission and subsequent spillover into human populations, which would better explain the spread and the bottlenecks observed in swine in different HEV epidemics. Overall, continued genomic surveillance of the HEV human and animal infection is required to assess adaptability and selection, which is increasingly important on the verge of an eventual vaccine deployment.
In conclusion, this study contributes to the hypothesis that humans are probably infected with HEV after contact with swine sources. This emphasizes the importance of checking the swine country of origin and improving sanitary control measures in order to prevent the spread of HEV infection in Italy.