1. Background
Chronic hepatitis C (CHC) is a major health care burden and a leading cause of end-stage liver disease and hepatocellular carcinoma (HCC) worldwide. Although non-uniform distributions of CHC in certain areas complicate the establishment of global and regional epidemiology, the global prevalence of CHC has been estimated as 2.35%, affecting approximately 160 million people (1).
In Taiwan, seroprevalences of hepatitis C virus (HCV) antibody positivity were estimated as 4.4% to 8.6% (2, 3). Upwards of 53% of CHC patients are infected with HCV genotype 1 (G1) (4). Either in most resource-limited areas worldwide or in most of the Asian countries, pegylated interferon (pegIFN) plus ribavirin (RBV) combination therapy remains the first-line standard of care (SOC) for CHC. The National Health Insurance program of Taiwan reimburses the cost of SOC for CHC. However, severe RBV-induced hemolytic anemia severely complicates CHC patients during SOC by causing suboptimal tolerance, poor compliance and early withdrawal from therapy (5). The risks of anemia have been shown to be higher in Asian than non-Asian patients (6). Concerns about ribavirin-induced anemia and related issues still remain central to patient.
Extensive accumulation of RBV in erythrocytes causes membrane oxidative damage, premature hemolysis and subsequent anemia (7). Anemia during CHC treatment was considered multifactorial. The haptoglobin phenotype, pretreatment platelet level (8), impaired renal function (9), high dose/body weight ratio, old age and female sex (10) have been previously demonstrated to be associated with anemia. Recent genome-wide association studies have advanced to a strong association between RBV-induced anemia and single-nucleotide polymorphisms (SNPs) in the inosine triphosphate pyrophosphatase (ITPA) gene on chromosome 20 in CHC patients on SOC (11).
Identifying patients at high risk of anemia is crucial for improving patient compliance and SOC outcomes for CHC patients (12). Novel treatment strategies or algorithms based on a combination of relevant pharmacogenetics and host factors may facilitate patient counseling prior to anemia events (13). However, limited studies included predictive modeling of severe anemia (hemoglobin, Hb < 10 g/dL) based on an index incorporating the strong predictor, ITPA SNP status (14, 15).
2. Objectives
Therefore, we estimated the effect of ITPA SNP status on severe anemia and treatment responses to construct a clinically practical predictive index for severe anemia during CHC combination therapy.
3. Patients and Methods
3.1. Ethics
The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Institutional Review Board of China Medical University Hospital. Informed consent was obtained from each patient included in the study.
3.2. Patients
From September 2005 to September 2013, consecutive East Asian patients with CHC G1 were screened and enrolled in a prospective cohort to analyze antiviral treatment responses. We defined CHC G1 as positive result for serum anti-HCV antibody (Abbott Laboratories, Abbott Park, IL, USA) for more than six months with detectable serum HCV G1 RNA (Cobas Amplicor HCV Monitor 2.0; Roche Diagnostics, Branchburg, NJ, USA). Patients with a history of any of the following conditions were excluded from the cohort; age < 20 years or > 75 years, treatment discontinued in less than four months after starting SOC with no severe anemia event, hepatitis B virus coinfection, human immunodeficiency virus coinfection, HCC, alcoholic liver disease, primary biliary cirrhosis, primary sclerosing cholangitis, Wilson’s disease, autoimmune hepatitis, hemochromatosis, previous IFN-based therapy, baseline Hb < 13 g/dL in men, Hb < 12 g/dL in women, nucleoside- or nucleotide-analog therapies other than RBV, decompensated cirrhosis and end-stage renal disease. Eligible patients were randomly assigned to the development and validation cohorts at a ratio of 4:1.
3.3. Combination Therapy
Patients received pegIFN αα-2a (Pegasys, Hoffmann-La Roche, Basel, Switzerland) at a dosage of 180 µg/week or pegIFN α-2b (Peg-Intron, Schering-Plough, Kenilworth, NJ, USA) at a dosage of 1.5 µg/kg/week subcutaneously. Oral RBV was prescribed for all patients at a daily dose of 1000 mg (body weight < 75 kg) or 1200 mg (body weight ≥ 75 kg) for 24 weeks or 48 weeks based on the baseline HCV RNA (< versus ≥ 6 log10 copies/mL) and virological response at week 4 (16, 17). Baseline and mean doses of RBV (mg/kg/day) on treatment were calculated for each patient. Erythropoietin (EPO) was considered if Hb fell below 10 g/dL during the treatment.
3.4. Treatment Monitoring
Blood biochemistry (Beckman Coulter, CA, USA) and complete blood count analyses (Sysmex HST-series, Kanogawa, Japan) were performed in the central laboratory at the medical center. HCV RNA load was quantified at baseline and monitored at weeks 4, 12, 24 and 48.
3.5. Clinical Endpoints
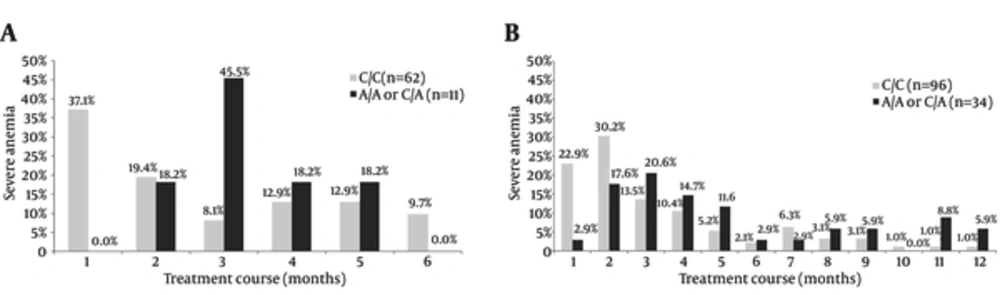
Severe anemia was defined as Hb level < 10 g/dL. Sustained virological response (SVR) was defined as undetectable HCV RNA at or before the end of treatment and at 24 weeks after the end of treatment. Monthly distributions of patients with severe anemia (hemoglobin < 10 g/dL) on treatment were compared between C/C versus “A/A or C/A” genotypes.
3.6. ITPA SNP Genotyping
Participant genomic DNA was extracted from peripheral blood mononuclear cells using a Qiagen DNA blood mini kit (Qiagen, Valencia, CA, USA). Genotyping for one functional missense ITPA variant in exon 2 at rs1127354 on chromosome 20 (18, 19) was performed using 20 µL of FastStart universal probe master mix (Roche Diagnostics, Branchburg, NJ, USA) containing 500 nmol rs1127354 forward primer (5’-TCTTGGAACAGGTCGTTCAGATTCTA-3’), 500 nmol rs1127354 reverse primer (5’-AGGAAGACAGAGAAATCCAACCATC-3’), 250 nmol C allele probe (FAM-AGTTTCCATGCACTTTGGTGG-BBQ) and 250 nmol A allele probe (YAK-AGTTTACATGCACTTTGGTGGC-BBQ). The reaction mixture was denatured at 95°C for 10 minutes before thermal cycling at 95°C for 15 seconds and 60°C for 1 minute for 40 cycles. Genotypes were analyzed using StepOneTM software Version 2.0 (Life Technologies, Carlsbad, CA, USA). Another reported ITPA SNP at rs7270101 is non-polymorphic in Asians. Therefore, patients were not genotyped for ITPA SNP rs7270101 (18). The cohort was also genotyped for interleukin 28B (IL28B) polymorphisms at rs8099917 and rs12979860, as previously described (20).
3.7. Statistical Analysis
Both Kolmogorov-Smirnov and Shapiro-Wilk tests showed that (P < 0.05) none of the continuous variables in this study had a normal distribution. Therefore, continuous variables were expressed as median (interquartile range, IQR) and estimated using Mann-Whitney U test. Categorical variables were estimated using the chi-square test or Fisher’s exact test. Univariate logistic regression identified the variables (P < 0.25) associated with severe anemia for subsequent multiple regressions. By expressing the likelihood of severe anemia as a probability ranging from 0 to 1, an index for identifying severe anemia was constructed by incorporating significant independent associated factors and the beta coefficients acquired through the final multiple logistic regression for severe anemia (14). The diagnostic performance of the predictive index was evaluated by the area under the receiver operating-characteristic curves (AUCs). The odds ratios (ORs) of significant associations were determined based on a 95% confidence interval (CI). The data was analyzed using SAS Version 9.3 (SAS Institute, Inc., Cary, NC, USA). A 2-sided P < 0.05 indicated statistical significance.
4. Results
4.1. Patient Characteristics
A total of 453 patients were screened. Excluded patients included those with baseline hepatitis B virus coinfection (n = 9), HCC (n = 5), a history of IFN-based therapy (n = 11), men with baseline Hb < 13 g/dL or women < 12 g/dL (n = 8) and end-stage renal disease (n = 2). Therefore, the CHC G1 cohort consisted of 418 eligible participants who intended to receive combination therapy. ITPA data were missing for four patients. Nine patients were also excluded, because the treatment was discontinued within four months from the treatment baseline for reasons other than severe anemia.
Thus, 405 patients entered the complete analysis. The cohort was randomized into a development cohort (n = 324) and a validation cohort (n = 81). Among them, 367 (90.6%) of 405 patients had liver histology data. A total of 148 (36.5%) patients received combination therapy for 24 weeks and 257 (63.5%) patients for 48 weeks. Patients per protocol (n = 357) entered the analysis for SVR (41 patients discontinued before the intended EOT and seven patients were lost to follow-up for SVR visit).
The median patient age was 53.0 (13.0) years and 49.6% of patients were men. The median baseline Hb was 14.4 (1.9) g/dL. The C/C genotype was present in 66.4% of patients and 33.6% had the A/A or the C/A allelic variants (“A/A or C/A” as a combo variable) at rs1127354. The development cohort (n = 324) and validation cohort (n = 81) did not differ significantly in a univariate analysis.
4.2. Predictive Index
Monthly distributions of severe anemia events are demonstrated in Figure 1. Among patients who received 24-week treatment and developed severe anemia, a significantly higher percentage of patients with C/C genotype did so during the first month on treatment than those with “A/A or C/A” genotype (37.1% versus 0.0%, P = 0.0135). Likewise, among patients who received 48-week treatment and developed severe anemia, a significantly higher percentage of patients with C/C genotype did so during the first month on treatment than those with “A/A or C/A” genotype (22.9% versus 2.9%, P = 0.0079). In total, 166 patients (166/324, 51.2%) in the development cohort developed severe anemia on treatment. Multiple logistic regressions identified age (≥ 50 y: OR = 9.7, 95% CI = 5.0 - 18.6), ITPA rs1127354 (C/C: OR = 3.3, 95% CI = 1.8 - 5.8) and baseline hemoglobin (< 14.0 g/dL: OR 6.4, 95% CI = 3.3 - 12.1; 14.0 - 14.9: OR = 2.4, 95% CI = 1.2 - 4.6) as predictors of severe anemia on treatment (Tables 1 and 2). There was no significant dose effect between ITPA SNP status (C/A versus A/A, P = 0.5387) and severe anemia. Therefore, the predictive index derived from the development cohort incorporating age (y), ITPA SNP status and baseline Hb (g/dL) was expressed as 1/(1 + exp (- x)), where x = - 3.2835 + 2.2709 × A + 1.1822 × I + 1.8848 × H1 + 0.8684 × H2 (A = 1 if age ≥ 50; A = 0 if age < 50; I = 1 if ITPA SNP = C/C; I = 0 if ITPA SNP = “A/A or C/A”; H1 = 1 if Hb < 14.0; H1 = 0 if Hb ≥ 14.0; H2 = 1 if Hb = 14.0 - 14.9; H2 = 0 if Hb < 14.0 or ≥ 15.0).
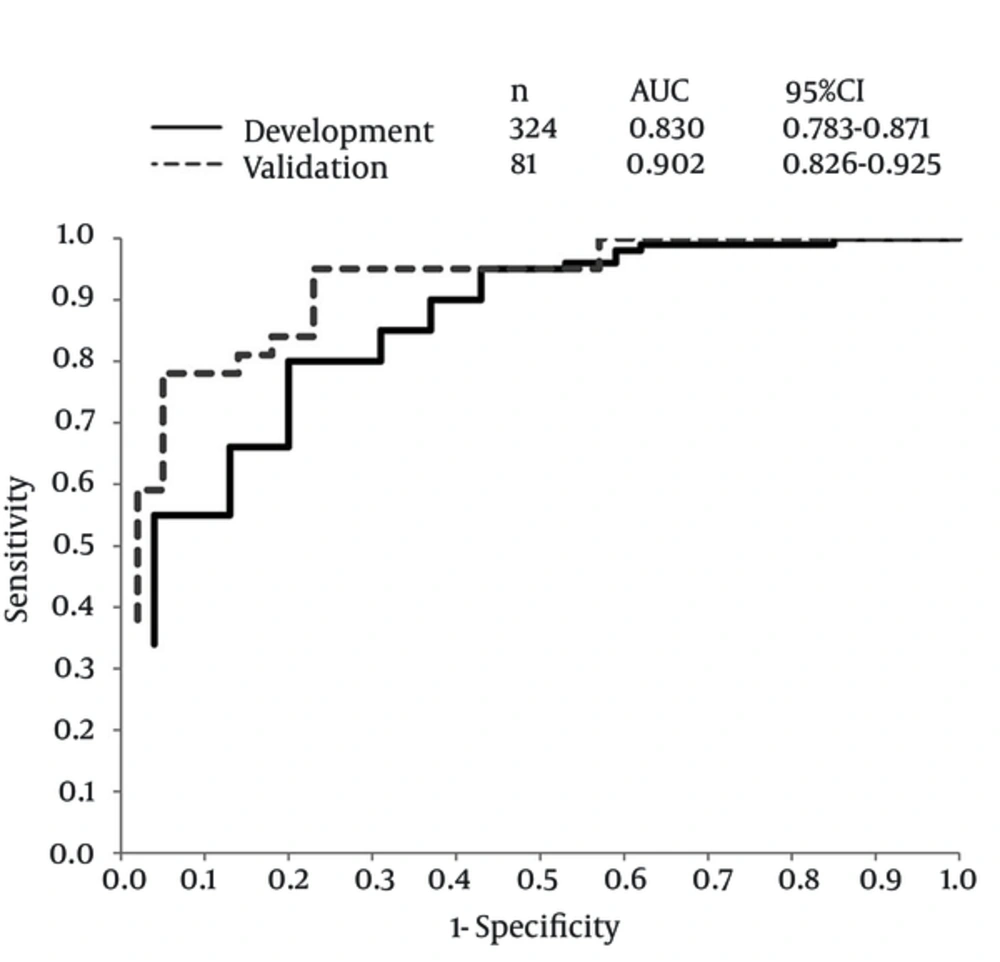
To dichotomize the anemia status (with versus without severe anemia) in the development, the cutoff value of 0.5 was optimal when maximizing the values of “sensitivity + specificity - 1” (21). The predictive index yielded a diagnostic accuracy (AUC) of 0.830 (95% CI = 0.783-0.871) in the development (n = 324) and 0.902 (0.826-0.925) in the validation (n = 81) cohorts to dichotomize the anemia status, respectively (Figure 2).
4.3. Treatment Responses
In total, 135 (135/357 per protocol, 37.8%) patients achieved RVR. Seventy-three of them (73/135, 54.1%) had a median baseline HCV RNA of 5.29 (0.76) log10 copies/mL and received 24-week treatment. The remaining 62 patients (62/135, 45.9%) received 48-week treatment. Liver pathology was acquired in 69 of 73 (94.5%) patients with RVR and 24-week treatment. Among them, METAVIR fibrosis stages 0 - 2 were noted in 54 (54/69, 78.3%), and 3 - 4 in 15 (15/69, 21.7%).
In total, 255 (255/357 per protocol, 71.4%) patients achieved SVR. The RVR (OR = 8.2; 95% CI = 3.5–19.1; P < .0001), T/T IL28B genotype at rs8099917 (OR = 3.0; 95% CI = 1.2 - 7.4; P = 0.0156), the METAVIR fibrosis stages 0-2 (OR = 1.9; 95% CI = 1.0 - 3.5; P = 0.0450), baseline HCV RNA (OR = 0.4; 95% CI = 0.2 - 0.7; P = 0.0008) and 48-week treatment duration (OR = 1.1; 95% CI = 1.0 - 1.1; P = 0.0003) were independently associated with SVR. Anemia events and EPO treatment were not independently associated with SVR. In patients with either ITPA C/C or “A/A or C/A” genotypes, the mean RBV dose exposures were not significantly different between patients with SVR versus non-SVR. The IL28B SNP (rs12979860) was not included in the final multiple regression because of collinearity with the other IL28B SNP (rs8099917) (Table 3).
In addition, through univariate analyses, the ITPA SNP status was significantly associated with RBV dose reduction in the development cohort (n = 324) (Mann-Whitney U test, P = 0.0035). Patients (n = 103/324, 31.8%) with “A/A or C/A” genotype received 100% (8%) intended RBV doses. In contrast, patients (n = 221/324, 68.2%) with C/C genotype received 97% (1.4%) intended RBV doses.
However, ITPA SNP status was not significantly associated with EPO use (chi-square test, P = 0.1617). Forty-five of 103 patients (45/103, 43.7%) with “A/A or C/A” genotype received EPO treatment. In contrast, 115 of 221 patients (115/221, 52.0%) with C/C genotype received EPO treatment.
A) Among patients who received 24-week treatment and developed severe anemia, a significantly higher percentage of patients with C/C genotype did so during the first month than those with “A/A or C/A” genotype (P = 0.0135). B); also, among patients who received 48-week treatment (P = 0.0079).
| Variables | Anemia (n = 166) | Non-Anemia (n = 158) | P Value |
|---|---|---|---|
| Age, y | < 0.0001 | ||
| < 50 | 17 (10.2) | 83 (52.5) | |
| ≥ 50 | 149 (89.8) | 75 (47.5) | |
| Gender | < 0.0001 | ||
| Female | 100 (60.2) | 56 (35.4) | |
| Male | 66 (39.8) | 102 (64.6) | |
| Body mass index, kg/m2 | 23.75 (3.8) | 25 (3.9) | 0.0011 |
| IL28B SNP (rs8099917) | 0.5189 | ||
| T/G or G/G | 24 (14.5) | 19 (12.0) | |
| T/T | 142 (85.5) | 139 (88.0) | |
| IL28B (rs12979860) | 0.5566 | ||
| C/T or T/T | 27 (16.3) | 22 (13.9) | |
| C/C | 139 (83.7) | 136 (86.1) | |
| ITPA (rs1127354) | 0.0002 | ||
| A/A or C/A | 37 (22.3) | 66 (41.8) | |
| C/C | 129 (77.7) | 92 (58.2) | |
| Platelet, 1000/µL | 155.0 (68.0) | 168.0 (82.0) | 0.0007 |
| Creatinine, mg/dL | 0.77 (0.30) | 0.81 (0.27) | 0.3375 |
| Baseline hemoglobin, g/dL | < 0.0001 | ||
| < 14 | 85 (51.2) | 32 (20.3) | |
| 14 - 14.9 | 47 (28.3) | 37 (23.4) | |
| ≥ 15 | 34 (20.5) | 89 (56.3) | |
| Alanine aminotransferase, IU/L | 85.0 (84.0) | 86.5 (74.0) | 0.3627 |
| METAVIR fibrosis stage | 0.4233 | ||
| 0 - 2 | 100 (69.9) | 109 (74.1) | |
| 3 - 4 | 43 (30.1) | 38 (25.9) | |
| HCV RNA, log10 copies/mL | 6.89 (0.94) | 6.89 (0.89) | 0.9754 |
| Interferon use | 0.2744 | ||
| < 100% intended dose | 27 (16.3) | 19 (12.0) | |
| Ribavirin dose, mg/kg/d | 14.9 (2.9) | 15.6 (2.1) | 0.0037 |
| Erythropoietin use | < 0.0001 | ||
| No | 47 (28.3) | 117 (74.1) | |
| Yes | 119 (71.7) | 41 (25.9) | |
| Treatment durations, week | 0.7906 | ||
| 24 | 54 (32.5) | 54 (34.2) | |
| 48 | 94 (56.6) | 84 (53.2) | |
| Discontinued | 18 (10.8) | 20 (12.7) |
a Data are presented as Median (IQR) or No. (%).
b Continuous variables were estimated using Mann-Whitney U test. Categorical variables were estimated using chi-square test.
| Variables | β Coefficient | Standard Error of β Coefficient | Odds Ratio (95% CI) | P Value |
|---|---|---|---|---|
| Age, y | < 0.0001 | |||
| < 50 (19 - 49) b | 1.000 | |||
| ≥ 50 (50 - 73) b | 2.2709 | 0.3325 | 9.688 (5.049,18.588) | |
| ITPA (rs1127354) | < 0.0001 | |||
| A/A or C/A | 1.000 | |||
| C/C | 1.1822 | 0.2977 | 3.262 (1.820, 5.845) | |
| Baseline hemoglobin, g/dL | < 0.0001 | |||
| < 14 (12.0 - 13.9) b | 1.8848 | 0.3308 | 6.352 (3.322, 12.147) | |
| 14-14.9 | 0.8684 | 0.3365 | 2.383 (1.232, 4.609) | 0.0099 |
| ≥ 15 (15.0 - 17.7) b | 1.000 |
a Data are presented for Hemoglobin < 10 g/dL and Development Cohort (n = 324).
b Range of variables, intercept = -3.2835.
| Variables | Univariate | Multiple | |||
|---|---|---|---|---|---|
| Sustained Virological Response | P Value | OR (95% CI) | P Value | ||
| (+) n = 255 | (-) n = 102 | ||||
| Age, y | 52.0 (14.0) | 55.5 (14) | 0.0296 | ||
| Gender | 0.8408 | ||||
| Female | 128 (50.2) | 50 (49.0) | |||
| Male | 127 (49.8) | 52 (51.0) | |||
| Body mass index, kg/m2 | 23.97 (4.0) | 25.04 (3.95) | 0.0110 | ||
| IL28B SNP (rs8099917) | < 0.0001 | 0.0156 | |||
| T/G or G/G | 17 (6.7) | 23 (22.5) | |||
| T/T | 238 (93.3) | 79 (77.5) | 3.032 (1.234 - 7.449) | ||
| IL28B SNP (rs12979860) | 0.0002 | ||||
| C/T or T/T | 21 (8.2) | 23 (22.5) | |||
| C/C | 234 (91.8) | 79 (77.5) | |||
| ITPA (rs1127354) | 0.9441 | ||||
| A/A or C/A | 89 (34.9) | 36 (35.3) | |||
| C/C | 166 (65.1) | 66 (64.7) | |||
| Platelet, 1000/µL | 169 (72) | 145 (71) | 0.0008 | ||
| Serum creatinine, mg/dL | 0.77 (0.3) | 0.8 (0.24) | 0.1199 | ||
| Hemoglobin, g/dL | 14.4 (2.0) | 14.4 (1.9) | 0.7231 | ||
| Alanine aminotransferase, IU/L | 85 (92) | 86.5 (66) | 0.7509 | ||
| METAVIR fibrosis stage | 0.0067 | 0.0450 | |||
| 0 - 2 | 174 (75.3) | 56 (60.2) | 1.886 (1.014 - 3.507) | ||
| 3 - 4 | 57 (24.7) | 37 (39.8) | 1.000 | ||
| HCV RNA, log10 copies/mL | 6.71 (1.14) | 7.02 (0.54) | < 0.0001 | 0.385 (0.220 - 0.673) | 0.0008 |
| Interferon use | 0.0068 | ||||
| < 100% intended dose | 27 (10.6) | 22 (21.6) | |||
| Ribavirin dose, mg/kg/d | 15.4 (2.6) | 14.85 (2.8) | 0.0461 | ||
| Erythropoietin use | 0.6388 | ||||
| No | 133 (52.2) | 56 (54.9) | |||
| Yes | 122 (47.8) | 46 (45.1) | |||
| Rapid virological response | < 0.0001 | < 0.0001 | |||
| No | 123 (49.4) | 84 (90.3) | 1.000 | ||
| Yes | 126 (50.6) | 9 (9.7) | 8.212 (3.535 - 19.079) | ||
| Treatment durations, wk | 0.0443 | 0.0003 | |||
| 24 | 86 (33.7) | 46 (45.1) | 1.000 | ||
| 48 | 169 (66.3) | 56 (54.9) | 1.051 (1.023 - 1.079) | ||
| Hemoglobin < 10, g/dL | 0.8935 | ||||
| No | 127 (49.8) | 50 (49.0) | |||
| Yes | 128 (50.2) | 52 (51.0) | |||
| Hemoglobin < 10, g/dL c | 0.8735 | ||||
| No | 226 (88.6) | 91 (89.2) | |||
| Yes | 29 (11.4) | 11 (10.8) | |||
| Hemoglobin decline ≥ 4, g/dL | 0.9166 | ||||
| No | 91 (35.7) | 37 (36.3) | |||
| Yes | 164 (64.3) | 65 (63.7) | |||
a Data are presented as Median (IQR) or No. (%).
b Continuous variables were estimated using Mann-Whitney U test. Categorical variables were estimated using chi-square test.
c Data are presented within 4 weeks.
5. Discussion
The frequency of severe anemia was 50.1% (203/405). This incidence was higher than that reported (39.3%) in a previous study of 466 East Asian patients diagnosed with CHC (5). The higher anemia frequency in our study can be attributed to the higher RBV dosage in patients infected with HCV G1. Consistent with Hb kinetics reported in previous studies (5, 12, 19, 22-25), the anemia events in our study accumulated during the first few months of treatment and remained relatively stable thereafter (Figure 1).
The accumulated ITP was substituted for GTP during the biosynthesis of ATP and protected against RBV-induced hemolytic anemia. Therefore, the wild C/C genotype for ITPA is hemolysis-susceptible. Methods of gene expression analysis such as mRNA transcripts or western blotting were not used in our study. However, the results of SNP genotyping of ITPA that we performed correlated closely with phenotypes or predicted ITPA activities of these allelic variants (26-28). Although RBV pharmacokinetics in serum or in erythrocytes or predicted ITPA activities were not applicable in our current study (29), this index may serve as a clinically practical one based on dichotomized parameters available in clinical settings. Future studies can develop algorithms or scoring systems based on age, sex, body weight, renal function, Hb and ITPA SNP status for modifying RBV dosage to achieve an optimal steady-state concentration range or for administering EPO prior to the development of anemia, rather than reducing the RBV dose following an anemia event (30, 31). Recent studies indicated that ITPA allelic variants “A/A or C/A” at rs1127354 are less likely associated with RBV dose reduction (19, 22, 23, 32). Future studies can also use a time-to-event analysis to identify significant predictors for RBV dose reduction.
The effects of older age and female sex on severe anemia (< 11 g/dL) during the first four weeks of treatment were also reported in a study of 61 HCV G1-infected Japanese patients, in which 49 patients exhibited RBV-sensitive ITPA C/C genotype at rs1127354 and 12 patients exhibited RBV-resistant “A/A or C/A” variants (19). Therefore, older female patients require stringent monitoring of anemia during combination therapy. However, sex was not included in the predictive index through multiple logistic regressions in the current study because of collinearity with baseline Hb. The serum creatinine levels did not gain significance during univariate analysis. Therefore, it was not applicable to adopt creatinine level in multiple regression modeling for predictive index of severe anemia. In the study by Tsubota et al., estimated glomerular filtration rate was found to be one of the significant associated factors to construct a predictive model for RBV-induced anemia (14). Our future studies would recruit participants in larger sample sizes receiving more uniform treatment durations to allow time-dependent analysis of anemia events for constructing a predictive index through Cox regression analysis.
Based on our results, we suggest that subjects with a predictive index greater than 0.5 might receive EPO treatment early after the start of pegIFN and RBV combination therapy to facilitate quality of life, prevent RBV dose reduction and avoid compromised treatment responses. However, anticipated benefits associated with early implementation of EPO treatment need to be confirmed by further studies.
Treatment responses are a major concern regarding SOC for CHC. Both RVR and T/T IL28B genotype at rs8099917 remained significant predictors for SVR in our CHC G1 cohort (33). Also consistent with recent studies (18, 19, 22-24, 34), ITPA SNP status was a non-significant predictor of SVR in our study. However, the reported ITPA-SVR correlations varied (35). In a recent report by Rembeck et al. (27), ITPA allelic variants encoding reduced ITPA activity was demonstrated to correlate with SVR and non-relapse, unrelated to RBV adherence or protection against anemia, although the molecular mechanisms require further elucidation.
Although previous reports yielded conflicting results regarding the significance of anemia-SVR correlation (5, 12, 36, 37), most recent studies on ITPA SNPs have not estimated anemia-SVR correlation (18, 19, 22-25, 32, 38). In our study, SVR was not associated with severe anemia or Hb decline (≥ 4 g/dL) on treatment (Table 3). Recent anemia-SVR correlations have also been reported based on the divergent results of non-genetic (5, 23, 36) and molecular studies (11, 14). Molecular studies indicated that anemia might not only be a surrogate for RBV exposure, because anemia-SVR association was attenuated when adjusting for RBV levels (39), but also represent the first molecular evidence connecting anemia-SVR association to novel genes and pathways through statistical analyses on gene expressions and mRNA signatures (11).
In conclusion, ITPA SNP status is a significant predictor of RBV-induced hemolytic anemia in patients diagnosed with CHC G1 and receiving pegIFN and RBV combination therapy. The ITPA SNP-based index is an accurate and practical predictive solution for severe anemia in clinical practice. Information on ITPA SNP status of patients may assist in maximizing the tolerability of SOC for patients with CHC.