1. Background
Hepatitis C Virus (HCV) is an infectious pathogen belonging to genus Hepacivirus and the Flaviviridae family that has infected more than 180 million people worldwide (1). Also, it was estimated that 1.75 million new HCV infections occurred worldwide in 2015; based on estimation, the number of individuals infected annually by HCV infection is estimated to be 3 to 4 million (2-4).
The most frequently affected regions are Mediterranean and European regions, with a prevalence of 2.3% and 1.5%, respectively (3, 5). Depending on the country, hepatitis C virus infection can be involved in certain populations and/or in general populations. There are multiple genotypes of HCV and their distribution vary based on region. Currently, the virus is classified to 7 genotypes and 67 subtypes with genotypes varying substantially in their geographical distribution (6). The most prevalent genotype is genotype 1 (subtypes 1a and 1b in the USA and Europe), followed by genotypes 3 (South Asia and Australasia), 2 (Sub-Saharan Africa), and 4 (Central Africa to the Middle East), respectively (7). Genotype 1 is predominant in non-Arab countries, including Turkey, Iran, and Cyprus (8).
In Iran, subtypes 1a, 3a, and 1b are the most prevalent subtypes (9). Hepatitis C prevalence in the general population of Iran is less than 0.5% (0.1% in females and 1.0% in males) (10). This distribution of HCV genotypes varies by region, as indicated by previous reports. These reports differed in 4 geographical regions of Iran, such that G1a and G3a were predominant genotypes in northern regions (11). Also, there are controversies regarding the correlation of HCV genotypes with any clinical outcome. A combination of different factors, such as the genotype, have been suggested to play a role in liver disease progression. Although the new therapy, Direct-Acting Antivirals (DAAs), is the current standard treatment for hepatitis C, this therapy type differs from one genotype to another, so that pre-therapy genotyping and prediction of liver damage severity is necessary (12).
2. Objectives
Considering that the data of HCV genotypes distribution can improve the current understanding of HCV epidemics in a region and that genotypes play a crucial role in assessing therapeutic decisions and approaches, therefore the present study aimed at determining the distribution of HCV genotypes in Rasht, Capital City of Guilan Province, Northern Part of Iran.
3. Patients and Methods
3.1. Study Population
All patients were registered at the Medical Clinical Centre of Rasht, a central part of Gilan province, between June 2014 and February 2015. The protocol of the present study was approved by the Ethics Committee of Gilan University of Medical Science. Consent forms were signed by all subjects. Also, a questionnaire consisting of demographic information, was completed for each patient, including clinical data, such as age, gender, Alanine Aminotransferase (ALT), HBsAg, and anti-HCV antibody status.
This study included 83 subjects with HCV infection. The inclusion criteria of this study were as follows: (a) age of more than 18 years old with the presence of anti-HCV antibodies (positive anti-HCV), (b) serum Alanine Aminotransferase (ALT) higher than the reference range, and (c) lack of treatment history for this illness. Also serum ALT levels in HCV-infected patients were higher than the reference range. All received samples were screened for HCV-RNA by RT-PCR. Negative HCV PCR and genotyping were excluded. Therefore, 81 RNA HCV positive samples for HCV genotyping were entered in the study.
3.2. Hepatitis C Virus RNA Isolation
Six milliliters of whole blood sample were collected from each individual and all plasma samples were isolated from each 0.5 M EDTA-treated blood samples by centrifuging at 3000 rpm for 15 minutes, and then were preserved at -80°C. According to the manufacturer's instructions, RNA was extracted from 200 μL of plasma, using the QIAamp viral RNA kit (Qiagen, Hilden, Germany).
3.3. Complementary DNA Synthesis
After extraction, viral RNA was resuspended in a total volume of 20 µL and reverse transcription was carried out with 200 U of Moloney murine reverse transcriptase, (Fermentas GmbH, Germany), random hexamer (0.5 µM), (Fermentas GmbH, Germany), 0.5 mM each of dideoxynucleotides (dNTPs), (Fermentase GmbH, Germany), and reverse transcriptase reaction buffer 5x for 1 hour at 37ºC, followed by an enzyme inactivation step at 95ºC for 15 minutes in a thermocycler machine (MJ Research, Watertown, MA, USA).
3.4. RNA Hepatitis C Virus-Polymerase Chain Reaction
The sequences of PCR primers, specific for 5’-UTR region of HCV genome, have already been published (13). The sequence was as follows: ExF; 5’-AGCGTCTAGCCATGGCGT-3’, ExR; 5’-GCACGGTCTACGAGACCT-3’, InF; 5’-GTGTCTGCGGAACCGG-3’, InR; 5’-GGGCACTCGCAAGCACCC-3’. The first round of PCR reaction mixture contained 5 μL of cDNA as template, 5 μL of 10x PCR buffer (Roche, Germany), 1 μL of 10mM dNTPs, 1 μL of each external primer and 0.3 μL of Taq DNA polymerase. The final volume was adjusted to 50 μL. The thermocycler machine (Eppendorf, Germany) was programmed at 94ºC for 5 minutes followed by 30 cycles of denaturation at 94ºC for 55 seconds, annealing temperature at 55ºC for 40 seconds, and extension at 72ºC for 40 second, and 10 minutes at 72ºC for final extension. In the second round (for nested PCR) 3 µL of the first round PCR product was used as template and 1 µL of each internal primer was added to the reaction. Other PCR components were similar to the first round except annealing temperature, which was 64°C for 40 seconds, and the final volume was adjusted to 50 µL. Finally, the PCR product was loaded onto 2% agarose gel, electrophoresed, and then visualized under a UV transilluminator. The PCR fragment was 174 bp.
3.5. Hepatitis C Virus Genotyping
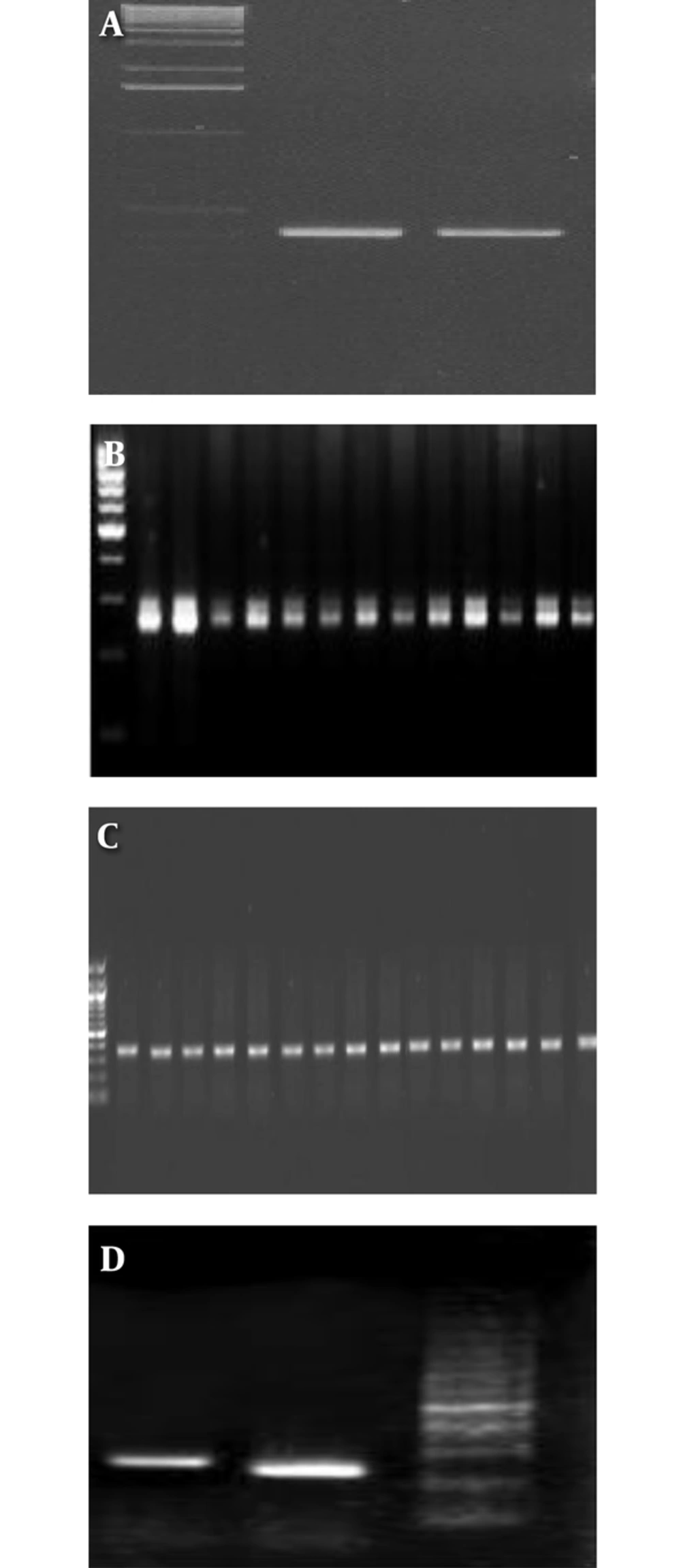
In this study, the PCR protocol was modified, and has previously been reported (14). Genotypes were determined by performing PCR, using specific primers for the target core region of the HCV genome (15). After cDNA synthesis, the PCR were performed. Briefly, cDNA synthesis and PCR amplification were performed in a 50-µL reaction volume, containing 20 µL of RNA, 50 pmol of each of primer, 200 µM of each deoxynucleotides (dNTPs), 10 U of Avian Myeloblastosis Virus Reverse Transcriptase (AMV RT) (Promega Madison, WI, USA), 2.5 U of Taq DNA polymerase (Promega Madison, WI, USA), 20 mM Tris-HCl (pH 8.4), and 50 mM KCl. Reactions were performed in a thermal cycler (Eppendorf, Germany), programmed as follows: 42°C for 30 minutes, 95°C for 5 minutes, 40 cycles at 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute, and 72°C for 5 minutes. Amplicons were then resolved by agarose gel electrophoresis (2%) with genotype-specific band sizes, which were compared with a 50-bp DNA ladder (Figure 1 A-D). Genotype 1a generates a 208-bp PCR product, genotype 1b, 2, and 3a generate 234, 337, and 285 bp PCR products, respectively. The plasma with different genotypes of HCV was used as a positive control for detection of different genotypes of HCV. Genotyping was confirmed by sequencing with gold standard methods, therefore, several samples were sent for sequencing.
3.6. Statistical Analysis
Analysis was performed by the SPSS software version 17.0, using descriptive values, such as mean, standard deviation, and Fisher’s exact probability test. T-test analysis was used for quantitative variables. P values of < 0.05 were considered statistically significant.
4. Result
Eighty-two patients with positive anti-HCV were recruited in this cross sectional study, out of which 81 were RNA HCV positive by PCR (Figure 2). The picture of the HCV genotyping products based on amplicon size is shown in Figure 1 A-D.
Demographic and HCV genotype distribution of all patients of this study are presented in Table 1. The mean age of patients was 49.6 ± 11.2 years (range: 19 to 62). The mean age of female participants was statistically higher than males (54.2 ± 3.8 versus 50.6 ± 4.6, respectively) (P < 0.05). The majority of the patients were aged 31 to 40 years, followed by 21 to 30 years.
| Variables | N | % | Mean ± SD |
|---|---|---|---|
| Age groups | 49.6 ± 11.2 | ||
| 21 - 30 | 34 | 42 | |
| 31 - 40 | 18 | 22.2 | |
| 41 - 50 | 17 | 21 | |
| ≥ 50 | 12 | 14.8 | |
| Gender | |||
| Male | 32 | 39.5 | |
| Female | 49 | 61.5 | |
| ALT (IU/L) | 41.5 ± 12.1 | ||
| AST (IU/L) | 27.3 ± 12.9 |
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase.
Hepatitis C Virus G1a was identified as the most abundant HCV subtype (42; 51.8 %). Furthermore, HCV G3a was observed in 23 participants (28.4%) followed by G2a, in 6 (7.4%), and G1b in 10 (12.4%). The HCV genotypes 5a and 6a were not detected in any of the HCV RNA positive patients. G1a was more frequent (85.7%) in male patients. The distribution of HCV genotypes in males was higher than females (68.3% versus 31.7%). G1a was the most frequent genotype in patients over 40 years of age (56.7% versus 41.2%) and G3a was the most frequent in patients under 40 years old (45.8% versus 38.3%), yet this difference was not significant. Also, the distribution of HCV genotypes based on other clinical data is shown in Table 2.
| Variables | GT3 (n = 23) | GT1a (n = 42) | GT2 (n = 6) | GT1b (n = 10) | P Value |
|---|---|---|---|---|---|
| Gender | 0.001 | ||||
| Male | 10 (43.5%) | 36 (85.7%) | 3 (50%) | 6 (60%) | |
| Female | 13 (56.5%) | 6 (14.3%) | 3 (50%) | 4 (40%) | |
| Age, mean ± SD | 43.1 ± 11.3 | 48.5 ± 1.8 | 46.1 ± 1.7 | 43.5 ± 4.7 | 0.32 |
| ALT | 23.3 ± 12.7 | 34 ± 11.7 | 29.4 ± 6.4 | 29.4 ± 3.4 | 0.56 |
| AST | 21.7 ± 11.8 | 28.6 ± 11.5 | 19.7 ± 11.8 | 18.6 ± 11.5 | 0.35 |
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase.
5. Discussion
Epidemiological studies in different regions and patients of the world show differences in distribution of HCV genotypes, therefore, the distribution of hepatitis C genotypes can contribute to health and treatment programs. Also, HCV genotype determination is an important need for clinicians in order to decide about the duration of antiviral treatment, because it is associated with the severity of liver damage and response to antiretroviral therapy (16). The geographical distribution of HCV genotypes is important for epidemiological studies in terms of distribution and possible risk groups. Therefore, it is necessary to determine the predominant genotype of the virus in each region, however, the aim of this study was to identify the prevalence of HCV genotypes in Rasht, Iran.
In the current study, HCV G1a was identified as the most abundant HCV genotype, which is the most prevalent worldwide and is the most frequently found in Northern Europe and North American (17). Previous reports showed that the most abundant HCV genotype was G1a (18), yet, in other studies conducted in Shiraz, Isfahan, and Mazandaran, G3a was the most common HCV genotype (19-21). The current results confirmed that the dominant genotype in Guilan province was 1a, which is in accordance with other reports from the same area of Iran. Based on previous studies in Guilan on HCV-infected patients and high risk individuals, such as hemodialysis and Beta-thalassemia, the most common genotype has been G1a (11, 22). Also, recent studies indicate that G1a 3a and 1b are the most prevalent genotypes in Iran (17). As shown previously, the distribution of HCV genotypes in Gilan in different studies has not changed, especially in high risk individuals; as these individuals have frequent blood transfusion and are at risk of exposure to infectious agents, such as HCV, this pattern may be due to low migration of different populations and the type of circulated HCV genotype. According to recent reports, a change in the distribution of HCV genotypes has been reported in a number of countries around the world (23-25). Therefore, the shift in the genotype circulation, especially in high risk individuals, may have a significant impact on clinical management of the patients as well as on the public health, therefore, this recent pattern can be concluding accurate clinical management of the patients.
In Iran, located in the Middle East, there has been regional reports on hepatitis genotyping among infected patients. Compared to the studies of Iran’s neighboring countries, it can be understood that the most common genotype is G1, found in approximately 50% to 75% of samples from Turkey and Bahrain (26-28). No study is available on HCV genotypic variation from Iran, Bahrain, and Pakistan, which could give cohesive circulation of HCV genotypes. Classification by viral genotype at the national and regional level, and a better understanding of viral diversity within different populations, will also critically inform the rational design and testing of HCV vaccines. Genotype diversity is particularly high in many Southeast Asian countries, and also in Western Europe, perhaps as a result of population immigration from Africa and/or Asia, therefore, the current results confirmed that it is better to investigate the distribution of HCV genotypes in each region of Iran based on periodic programs (29).
Male gender was reported as an independent predictive factor for HCV infection in previous studies (30). In this study, it was indicated that the difference of distribution of HCV genotypes between males and females was statistically significant. This may be because most of the HCV infected patients were male. Thus, it was not possible to analyze the distribution of HCV genotypes, according to gender, because of the low sample size. Similar studies have shown that the prevalence of hepatitis C in the general population of Iran is less than 0.5% (0.1% in females and 1.0% in males) (10); also, the difference in the distribution of HCV genotypes between males and females in HCV patients was statistically significant (31-33). In another report, the distribution of HCV G1 was found to be significantly associated with male gender, while G4 has frequently found in females (34). Also, in other studies, it was shown that there is no difference between both genders (35).
Epidemiological differences in age distribution of the HCV genotype have been previously investigated (17). The current results showed that distribution of HCV genotypes did not vary with age. European studies show that G1a and G3a are relatively more common in young individuals. In some studies, G1b-infected patients were significantly older than those infected by other genotypes and a significant proportion of younger patients were infected with G3 (21). In some studies, it was determined that G1a and G1b were more common in people older than 50 years of age, whereas G1b was the most common subtype among people younger than 20 years old (36). Hepatitis C virus increased in older patients compared with younger individuals, whereas there was no significant difference between males and females, which is incompatible with the current findings. In addition, according to a previous study from Iran, the prevalence of HCV infection in males was higher than females.
In conclusion, in the present study, G1a was the most prevalent genotype in Rasht, Iran. A high rate of HCV infection was reported in male patients. Further information is required on the epidemiology of HCV genotypes, as response to treatment is reported to vary in different types of HCV. This study had some limitations, such as subtype, with a relatively small sample size, and clinical data, such as therapy, yet the results may be helpful for understanding the genotype distribution of HCV in Rasht and can provide important information about HCV prevalence among different ages and gender.