1. Background
Chronic infection with hepatitis B virus (HBV) remains a serious public health problem worldwide. The biomarkers of HBV infection are important for disease prevention, diagnosis, and treatment management. The mostly applicable biomarkers of HBV infection include serum hepatitis B surface antigen (HBsAg), hepatitis e antigen (HBeAg), several antibodies against different HBV antigens, and HBV DNA levels (1). In addition, the circulating HBV RNA was suggested as a new biomarker in the latest EASL clinical practice guidelines (2).
HBV is known as a unique DNA virus with a reverse transcription step during replication cycle. The pregenomic RNA (pgRNA) is an important intermediate molecule connecting the parental and offspring HBV DNA. Theoretically, any host or viral factors that increase the production of pgRNA or block the reverse transcription of pgRNA in relaxed circular DNA may cause excess of pgRNA. Accumulating evidences suggest that excess pgRNA might present in virion-like particles and represent the majority of detectable HBV RNA in sera (3) and supernatant of HBV cell cultures (4, 5).
The serum HBV RNA are reported detectable in patients with chronic HBV infection (3, 6, 7) and its clinical significance are also suggested (8-10). It showed that serum HBV RNA dynamics had a strong correlation with HBeAg loss in patients treated with nucleoside-analogue or peginterferon (PegIFN)-α (9). It also showed that serum HBV RNA level is a valuable early predictor for the occurrence of antiviral resistant mutations (10) and reactivation of chronic hepatitis B (CHB) after antiviral therapy cessation (8). Serum HBV RNA is a novel biomarker of HBV infection and is useful to optimize antiviral strategies (9, 11). However, to understand the factors affecting its serum level and detection, and its biological and clinical significance, large cohorts of untreated patients are required.
2. Objectives
The current study aimed at analyzing serum HBV RNA in 483 patients with untreated HBeAg-positive and -negative CHB genotype B or C infection, the most prevalent HBV genotypes in China and Asia (12, 13). The study also aimed at finding the potential factors affecting the levels of serum HBV RNA and revealing how it is correlated with the other viral factors, such as serum HBV DNA and HBsAg. The study also aimed at giving a better understanding of this potential new biomarker for HBV infection.
3. Methods
3.1. Patients
Serum samples were provided from 483 patients with untreated CHB recruited in the current study. They were from registered and ethically approved clinical trials (NCT01088009 (13) and Institutional Review Board of Peking University: IRB00001052-14005). Each patient signed the informed consent. The diagnosis of CHB was according to the 2010 guidelines of prevention and treatment for chronic hepatitis B in China (14). None of the patients had autoimmune hepatitis, hepatitis C virus (HCV), hepatitis D virus (HDV), or human immunodeficiency virus (HIV) coinfection, primary biliary cirrhosis, and alcohol or drug abuse.
The 483 blood samples were collected from March 2010 to October 2015 (326 samples collected in 2010, 38 in 2011, 52 in 2012, 10 in 2013, 34 in 2014, and 23 in 2015). The HBeAg-negative samples were all collected from January 2014 to September 2015. They were properly kept at -80°C in aliquots. To ensure the stability of serum HBV RNA in sera, a 6-round repeated freezing (at -80°C) and thawing (at room temperature) experiment for 3 randomly selected serum samples were carried out before the pilot experiment and results showed no significant impacts of the treatments on HBV RNA levels (Appendix 1).
The serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were measured as previously described (13).
3.2. Serological Tests for HBV Marker
The serum HBsAg and HBeAg were detected by chemiluminescent microparticle immunoassay using ARCHITECT i2000 analyzer (Abbott Diagnostics, North Chicago, IL). The quantification of serum HBsAg was carried out by ARCHITECT HBsAg kit (Abbott Diagnostics) with a detection range of 0.05 - 250 IU/mL.
3.3. Measurement of Serum HBV DNA Level and genotyping
The serum HBV DNA level was quantified by TaqMan® 48 automatic florescence quantitative polymerase chain reaction (q-PCR) kits using Roche COBAS® AmpliPrep®/COBAS® TaqMan® 48 Analyzer (Roche Diagnostics, Mannheim, Germany); the detecting limit was 12 IU/mL.
The HBV subgenotyping by reverse transcriptase (RT) sequencing was performed following the standard operating procedure of the laboratory as described in authors’ previous reports (12, 15). The phylogenetic trees construction and nucleotide divergence analyses with clinically isolated and reference sequences for known subgenotypes were carried out by MEGA 7.0 software using neighbor-joining method and Kimura 2-parameter algorithm with bootstrapping 1000 replicates. Twenty-four subgenotype reference sequences were extracted from GenBank.
3.4. Measurement of Serum HBV RNA Level
The procedure of serum HBV RNA quantification was carefully described in the authors’ previous paper published in 2017 (16). The procedure included serum HBV nucleic acid extraction, DNA digestion, RT of RNA to complementary DNA, and real-time PCR. Due to the limitation of word counts, the readers are referred to the reference 16 or Appendix 2, patients and methods to this paper, for the detailed information. The lower limit of detection (LoD) for serum HBV RNA was 66.7 IU/mL (1.8 log IU/mL).
3.5. Statistical Analyses
Statistical analyses were performed with SPSS version 20.0 (SPSS Inc., Chicago, USA). The Student t test as well as nonparametric and Chi-squared tests was used to evaluate the appropriateness of the data. Logistic regression analysis was used to test the demographic characteristics as well as virological and biochemical factors that might influence the detectability of serum HBV RNA. Correlation analysis was used to study the relationship of serum HBV RNA with transaminases, HBV DNA, and HBsAg levels, in which the LoD values were used for those reported lower than LoD or undetermined. P < 0.05 was considered as statistically significant.
4. Results
4.1. Study Population
Of the 483 patients with CHB, 426 were HBeAg-positive and 57 HBeAg-negative (Table 1). The patients were infected with either genotype B or C CHB.
| Characteristic | HBeAg-Positive | HBeAg-Negative |
|---|---|---|
| Case number | 426 | 57 |
| Age, y, mean (range) | 30.0 (18.0 - 64.0) | 51.0 (19.0 - 69.0) |
| Gender, male/female (male%) | 327/99 (76.8) | 39/18 (68.4) |
| ALT (U/L), mean (range) | 123.6 (40.0 - 1150.0) | 82.0 (11.0 - 1730.0) |
| AST (U/L), mean (range) | 72.0 (25.0 - 985.0) | 49.0 (20.0 - 998.0) |
| HBV genotype, B/C (B%) | 153/273 (35.9) | 9/48 (15.8) |
| HBV DNA, log IU/mL, mean (range) | 8.0 (4.0 - 9.8) | 5.5 (3.2 - 7.8) |
| HBsAg, log IU/mL, mean (range) | 4.2 (0.8 - 5.5) | 2.4 (0.3 - 3.9) |
Abbreviations: ALT, Alanine Aminotransferase; AST, Aspartate Transaminase; HBsAg, Hepatitis B Surface Antigen; HBV, Hepatitis B Virus.
4.2. Detection of Serum HBV RNA
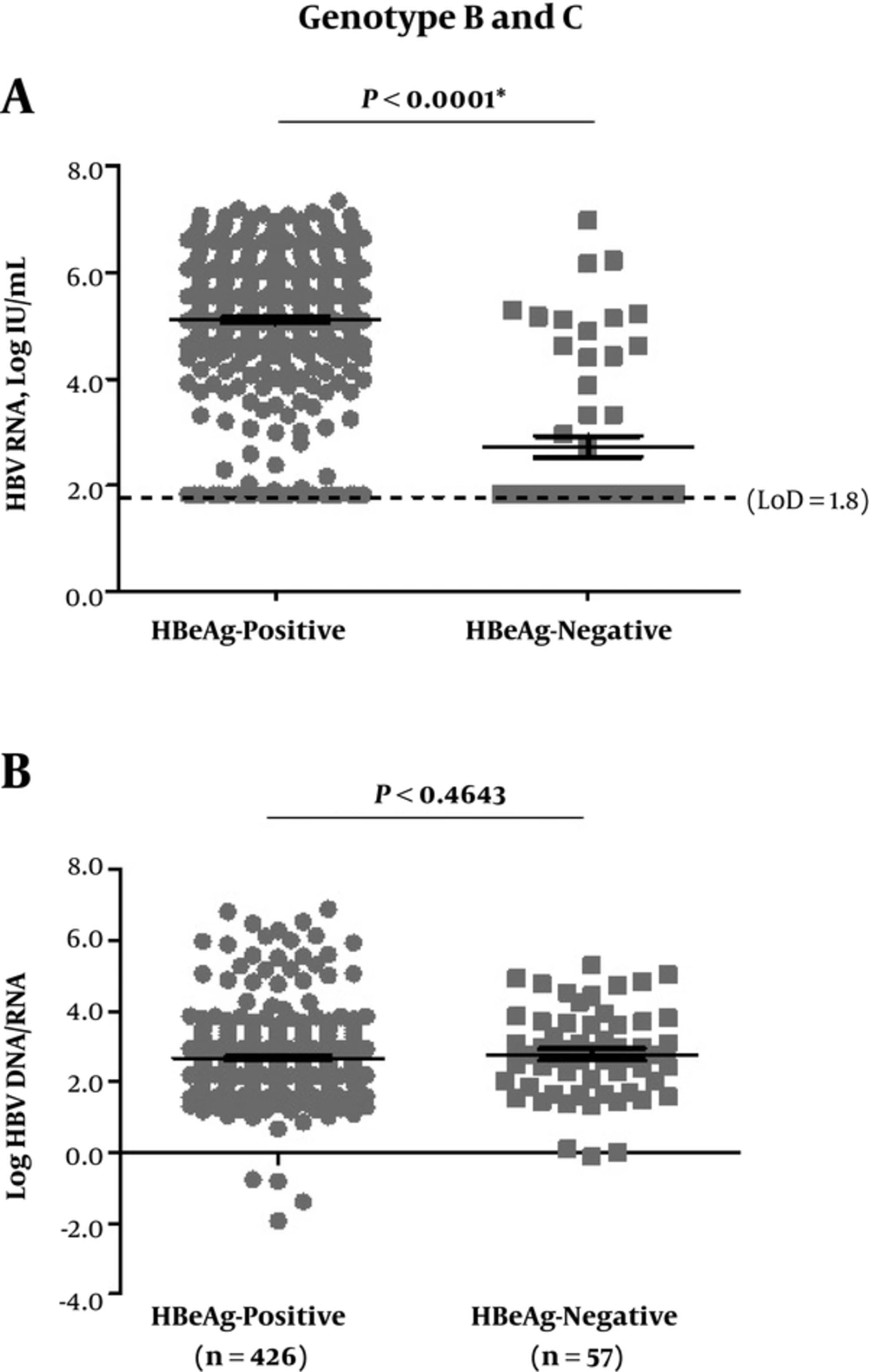
As shown in Figure 1A, serum HBV RNA was detected in 92.5% (394/426) of the patients with HBeAg-positive and 31.6% (18/57) with HBeAg-negative (P < 0.0001); the mean level was 5.4 (ranged: 1.8 - 7.3) log IU/mL in the ones with HBeAg-positive and 1.8 (ranged: 1.8 - 7.0) log IU/mL in HBeAg-negative ones (P < 0.0001). However, the logarithmic values of HBV DNA to HBV RNA ratio (log HBV DNA/RNA) showed no significant difference between HBeAg-positive and -negative cases (Figure 1B).
A, HBV RNA levels; B, log HBV DNA/RNA. The dotted line represented LoD of serum HBV RNA detection (66.7 IU/mL, 1.8 log IU/mL). The statistics here included samples with testing values reported lower than LoD or undetermined, and LoD values were used for these samples. CHB, chronic hepatitis B; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; LoD, lower limit of detection.
4.3. Potential Impact Factors of Serum HBV RNA Detection
Based on the data shown in Table 2, serum ALT and HBV DNA levels, as well as genotype were the independent predictors for serum HBV RNA detectability by the regression analysis in patients with HBeAg-positive (P < 0.05). The serum HBsAg level was found as an independent predictor using univariate analysis (P = 0.000), and was not excluded in multivariate analysis (P = 0.074).
| Factors | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P Value | OR | 95% CI | P Value | |
| HBeAg-positive, n = 426 | ||||||
| Age, y | 0.963 | 0.930 - 0.997 | 0.035a | |||
| Gender (male) | 1.110 | 0.482 - 2.555 | 0.806 | |||
| ALT, U/L | 1.004 | 0.999 - 1.008 | 0.092 | 1.005 | 1.001 - 1.010 | 0.024a |
| AST, U/L | 1.004 | 0.998 - 1.010 | 0.204 | |||
| HBV genotype, B | 4.257 | 1.464 - 12.377 | 0.008a | 4.052 | 1.321 - 12.428 | 0.014a |
| HBV DNA, log IU/mL | 2.506 | 1.807 - 3.474 | 0.000a | 2.126 | 1.352 - 3.343 | 0.001a |
| HBsAg, log IU/mL | 3.258 | 2.056 - 5.162 | 0.000a | 1.768 | 0.946 - 3.302 | 0.074 |
| HBeAg-negative, n = 57 | ||||||
| Age, y | 0.975 | 0.927 - 1.026 | 0.336 | |||
| Gender (male) | 0.899 | 0.270 - 2.930 | 0.847 | |||
| ALT, U/L | 1.004 | 1.000 - 1.007 | 0.046a | |||
| AST, U/L | 1.005 | 1.000 - 1.010 | 0.042a | |||
| HBV genotype, B | 1.750 | 0.325 - 9.410 | 0.514 | |||
| HBV DNA, log IU/mL | 5.142 | 2.079 - 12.718 | 0.000a | 5.142 | 2.079 - 12.718 | 0.000a |
| HBsAg, log IU/mL | 0.977 | 0.666 - 1.433 | 0.906 | |||
Abbreviations: ALT, Alanine Aminotransferase; AST, Aspartate Transaminase; CI, Confidence Interval; HBsAg, Hepatitis B Surface Antigen; HBV, Hepatitis B Virus; OR, Odds Ratio
aP < 0.05.
In patients with HBeAg-negative, the regression analysis revealed that serum HBV DNA was the independent predictor (P = 0.000). Although ALT and AST were also found as predictors in univariate analysis (P = 0.000), they were not confirmed in multivariate analyses.
4.4. Correlation Analysis of Serum HBV RNA Levels with Liver Transaminases, HBV DNA, and HBsAg Levels
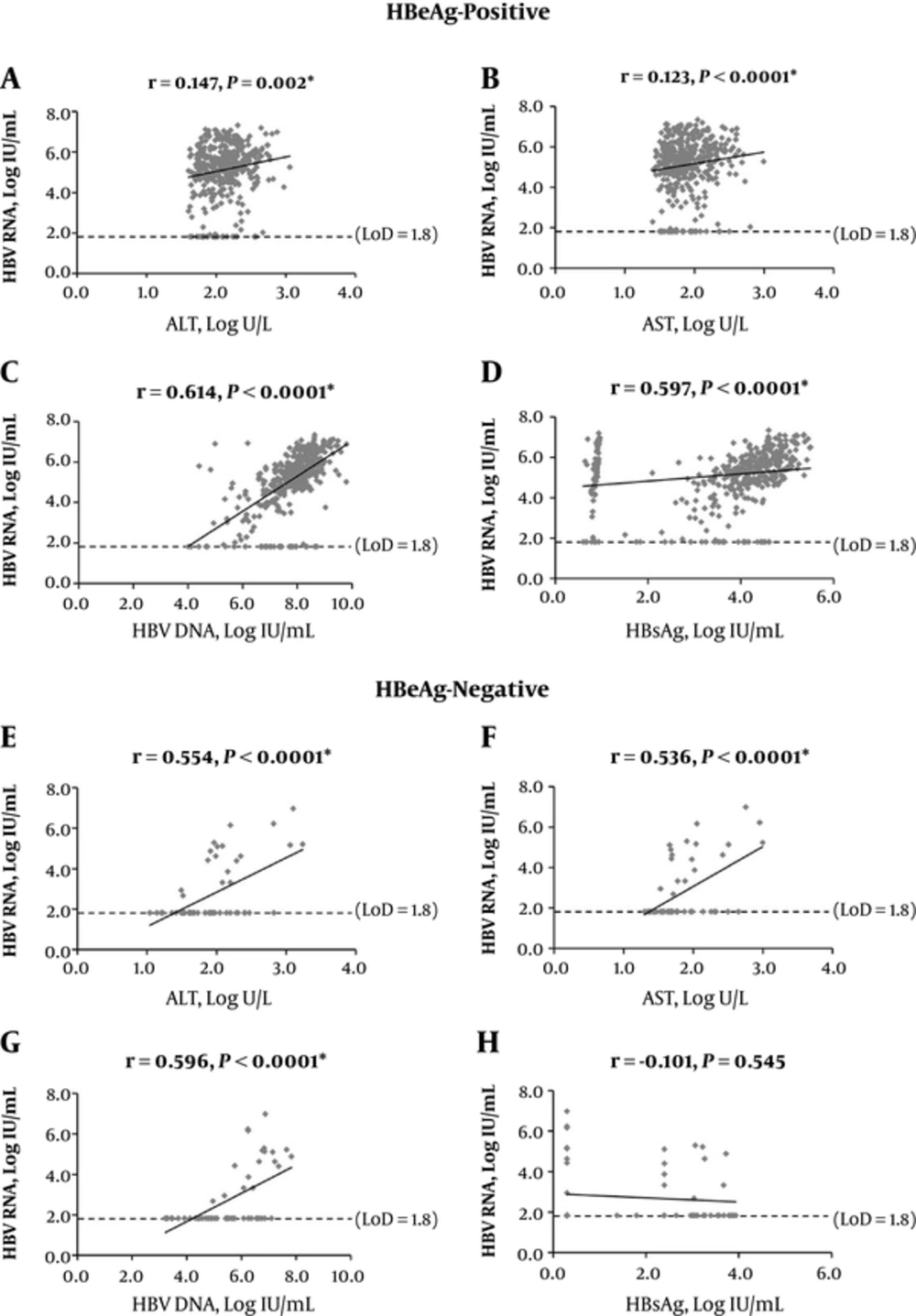
The results showed a significant positive correlation of serum HBV RNA with ALT, AST, and DNA levels (P < 0.05), regardless of HBeAg status (Figure 2). Positive correlations with HBsAg were only found in HBeAg-positive cases (P < 0.0001) (Figure 2D), and not the negative ones (P = 0.454) (Figure 2H).
A-D, HBeAg-positive; E-H, HBeAg-negative. The dotted line represented LoD of serum HBV RNA detection (66.7 IU/mL, 1.8 log IU/mL). The statistics here included samples with testing values reported as lower than LoD or undetermined, and LoD values were used for these samples. ALT, alanine aminotransferase; AST, aspartate transaminase; CHB, chronic hepatitis B; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; LoD, lower limit of detection.
4.5. Comparison Between Genotype B and C in Patients with HBeAg-Positive CHB Infection
Aforementioned results revealed HBV genotype and HBeAg statuses as the important factors affecting serum HBV RNA detection. However, the distribution of HBV genotypes B and C amongst the 2 groups of HBeAg-positive and -negative samples was not homogeneous. Thus, the confounding factors might affect the data interpretation.
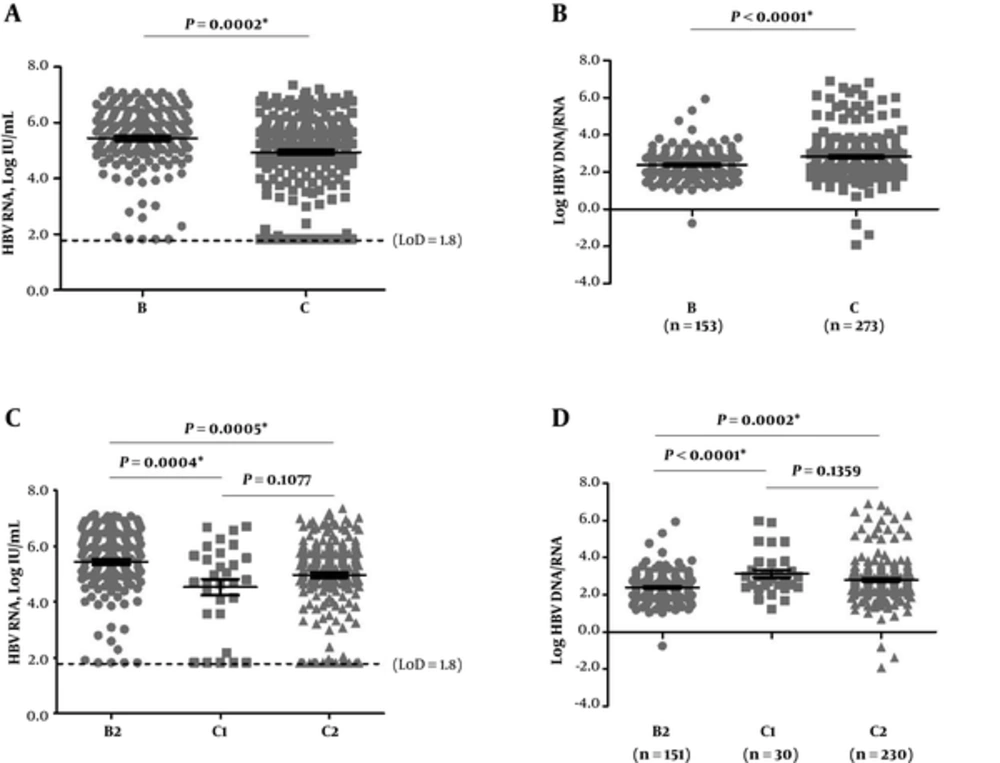
To further verify the current study findings, first, the serum HBV RNA was compared between genotype B and C in patients with HBeAg-positive CHB infection. Patients with HBeAg-negative CHB genotype B infection had very small sample size (n = 9) to be statistically powerful and were not included in the analyses. Based on Figure 3A, serum HBV RNA had significantly higher detectable rates and levels (149/153, 97.4%; mean (range): 5.6 (1.8 - 7.1) log IU/mL) in genotype B compared with genotype C (245/273, 89.7%; 5.3 (1.8 - 7.3) log IU/mL) (P = 0.0074; 0.0002). However, genotype C infection had significantly higher log HBV DNA/RNA (mean (range): 2.7 (-1.9 - 6.9)) than genotype B infection (2.3 (-0.8 - 5.9)) (P < 0.0001) (Figure 3B), though the serum HBV DNA levels had no significant difference between the genotypes (mean (range) B vs. C: 8.0 (4.7-9.8) vs. 7.9 (4.0-9.8) log IU/mL) (P = 0.8597).
A and C, HBV RNA levels; B and D, log HBV DNA/RNA. The dotted line represented LoD of serum HBV RNA detection (66.7 IU/mL, 1.8 log IU/mL). The statistics here included samples with testing values reported lower than LoD or undetermined, and LoD values were used for these samples. HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; LoD, lower limit of detection
Subgenotypes might also affect the results. Thus, HBV subgenotypes was further analyzed and identified 3 subgenotypes; i e, 33.1% (160/483) B2, 6.6% (32/483) C1, and 57.1% (276/483) C2. The subgenotypes of 15 sequences (2 B and 13 C) were undefinable. The B2, C1, and C2 accounted for 96.9% (468/483) of the sequences and used for the following subgenotype-based analysis. The results (Figure 3C and D) confirmed genotype-based findings (Figure 3A, B).
4.6. Comparison Between Patients with HBeAg-Positive and -Negative CHB Genotype C Infection
The current study compared serum HBV RNA levels between patients with HBeAg-positive and -negative CHB genotype C infection in order to verify the impact of HBeAg status on serum HBV RNA levels. Based on the data provided in Appendix 3 A, serum HBV RNA detectable rates and levels were significantly higher in patients with HBeAg-positive (245/273, 89.7%; mean (range): 5.3 (1.8 - 7.3) log IU/mL) compared with HBeAg-negative ones (16/48, 33.3%; 1.8 (1.8 - 7.0) log IU/mL) with CHB genotype C infection (P = 0.0008; < 0.0001). However, there was no significant difference in the log HBV DNA/RNA ratio between the groups (P = 0.8740) (Appendix 3 B). the current study results also confirmed the findings in subgenotype C2 (Appendix 3, C and D).
4.7. Evolutionary Analysis and GenBank Accession Numbers
The present study provided the phylogenetic trees (Appendix 4 and 5) and nucleotide divergence data (Appendix 6 and 7) for the sequences with defined subgenotypes by the evolutionary analysis. All the sequences obtained in the current study were also submitted to GenBank with accession numbers from MG714927 to MG715409.
5. Discussion
The current study found an 85.3% (412/483) detection rate for serum HBV RNA in the studied patients. However, 2 other reports by Huang et al., showed that serum HBV RNA was undetectable in all the 19 studied patients and only found in 40% (21/52) of the untreated ones (11, 17). The difference between the results of the current study and those of others may be attributed to the differences between the studied populations. The patients in the current study had a significantly higher HBeAg-positive ratio (426/483, 88.2%) compared with their patients (33/71, 46.5%) (P < 0.0001). The present study also showed that the ones with HBeAg-negative had a lower serum HBV RNA detection rate compared with the HBeAg-positive ones (P < 0.0001). After excluding the confounding factor of HBV genotype, the impacts of HBeAg status on serum HBV RNA level in genotype C and subgenotype C2 were also confirmed (Appendix 3). It is evident that HBeAg level might be affected by HBV precore G1896A mutation and core promoter A1762T/G1764A mutations (18, 19); A1762T/G1764A mutations were also reported to enhance viral replication in vitro. Patients with HBeAg-negative were more prone to such mutations compared with HBeAg-positive ones. Although the difference of serum HBV RNA detection between the patients with HBeAg-positive and -negative can be attributed to such mutations, further studies on patients with different HBeAg statuses and genotypes may better reveal the underlying mechanisms.
In addition to HBeAg status, the current study indicated that serum HBV DNA level was the only virological factor showing statistically coherent impact on serum HBV RNA level in both groups of patients with HBeAg-positive and -negative CHB infection. Positive correlations were also observed between serum HBV DNA and RNA levels in the study groups. However, the detection of serum HBV DNA could not guarantee the detection of serum HBV RNA, suggesting their intrinsic relationship, but different properties. Results of the current study suggested that the HBeAg status and serum HBV DNA level are the important viral factors affecting serum HBV RNA. Hence, it is necessary to characterize serum HBV RNA during the natural history of HBV infection in order to better evaluate its usefulness as a new biomarker of HBV infection in the clinical settings.
The origins of serum HBV RNA are still under investigation. The current speculations include the immature RNA-containing virion as a byproduct of normal HBV replication and exosome secretion (20). The current study data showed that the serum HBV RNA level was positively correlated with serum ALT and AST levels (Figure 2). This association was more obvious for ALT in patients with HBeAg-positive (Table 2). The high ALT and AST levels were the indicators of hepatocyte injury by human immune responses (21). In this scenario, HBV replication intermediates containing both HBV DNA and RNA are released from the injured hepatocytes. Results of the present study revealed the effect of hepatocyte injury on HBV RNA release. The detection of serum HBV RNA might reflect the outcome of complex viral and host interactions. More investigations are needed to fully understand this.
HBV genotype was identified as another important factor affecting serum HBV RNA level in patients with HBeAg-positive (Table 2). The finding was also verified in the subgenotype B2, C1, and C2 (Figure 3). It has long been known that different genotypes had marked differences in HBV natural history, disease progression, treatment response, and in vitro replication efficacy (22, 23). In terms of genotype differences, the authors’ previous study found that the core promoter transcriptional activity of genotype B was significantly higher than that of genotype C, hinting more production of pgRNA in genotype B (19). Similarly, the log HBV DNA/RNA of genotype B was significantly lower than that of genotype C in the present study (Figure 3B). The genomic makeup of HBV and subgenotype-related mutations might affect the viral DNA, RNA, and protein production.
Interestingly, the serum HBV RNA was never considered as it is today. Studies suggested that it might be a significant clinical novel biomarker (5, 8, 9). Halgand et al., showed a correlation between the detection of pgRNA and cccDNA in tumors, and the absence of tumorous microvascular invasion and better patient survival (24). Wang et al., revealed that the serum HBV RNA level is in correlation with the intrahepatic HBV RNA level, intrahepatic HBV RNA, cccDNA, and the histological scores for grading and staging (25). The current study also found that the pretreatment intrahepatic cccDNA was positively correlated with serum HBV RNA (r = 0.25, P = 0.02) (16). The accumulating evidence suggests that the serum HBV RNA level might reflect the intrinsic characteristic of HBV replication resulting from the net efficiencies of pgRNA production, reverse transcription, and hepatocyte injury as well.
In conclusion, the current study found that the existence of serum HBV RNA can be a general virologic marker for patients with HBeAg-positive CHB infection, but not the HBeAg-negative ones during the natural history of HBV infection. The serum HBV RNA level could be affected by HBeAg status, serum HBV DNA, HBsAg levels, HBV genotype, and ALT and AST levels. As a new biomarker of HBV infection, it might have similar, but yet distinct, characteristics as serum HBV DNA and HBsAg. Its roles in viral replication, infection, survival, disease progression, and antiviral response should be investigated in further studies.