1. Background
Hepatitis B virus (HBV) infections are a serious threat to human health. Over two billion people worldwide have been infected with HBV (1), and approximately 600,000 people die of HBV-related diseases each year (2).
China is home to an estimated 9300 million people living with HBV, among whom approximately 20 million are chronic cases, resulting in 300,000 deaths each year (3). Vaccination is an effective means of protecting individuals from HBV infection. However, some patients were infected with HBV before the hepatitis B vaccine became widespread or were not sufficiently protected by the vaccine (4). These patients may suffer from serious HBV-related diseases and new disease phenotypes may arise.
As of now, 10 distinct HBV genotypes (A to J) have been identified, representing 8% of whole-genome sequence divergence (5-7). The distribution and prevalence of HBV genotypes have regional characteristics (8-10) and influence the progression of liver-associated diseases (11-13). The occurrence of HBeAg-negative chronic hepatitis (CHB) is related to specific genotypes (14). Among two groups of Mexicans, patients with liver disease were found to have HBV genotype D, while homosexuals only had HBV genotype G (15). Furthermore, HBV genotype C is associated with a more efficient immuneresponse and an earlier hepatitis B e antigen (HBeAg) seroconversion, yet it can induce severe necro-inflammation (16, 17) and confers a higher risk for developing HCC than genotype B in East Asians (18). In South Africa, the risk of developing liver cancer among patients exposed to sub-genotype A1 is 4.5 folds higher than those exposed to other genotypes (19). HBV genotype I has recently been identified in northeast India, Laos, Vietnam, and China (5, 20-23). Although this particular genotype has a wide distribution, in-depth studies of this specific genotype are limited.
The coexistence of HBsAg and anti-HBs is a special serological state, with a rate of occurrence of 2.43% to 8.9% in different areas. This may be associated with immune evasion and HBV-related liver diseases (24-26). An increase in the incidence of HBsAg alongside anti-HBs has recently been reported. The mechanism underlying the phenotype in CHB patients remains unclear, and the selection for HBsAg variants that evade the immune system might be one possible reason for this.
2. Objectives
The current study identified a HBV strain isolated from a CHB patient from Zhaotong city, Yunnan province, China, who was infected with HBV genotype I and presented HBsAg and anti-HBs coexistence. In an effort to elucidate the mechanism underlying the coexistence of HBsAg and anti-HBs in this patient, full-length genome amplification was performed, and the HBV strain was cloned to conduct a comprehensive analysis of mutations and to analyze quasi species variation in relation to the coexistence of HBsAg and anti-HBs.
3. Methods
An original study of the full-length genome of HBV quasi species was performed alongside a meta-analysis to elucidate the mechanism underlying the coexistence of HBsAg and anti-HBs in a CHB patient with genotype I.
3.1. Patient
An HBV-infected, treatment-naive 58-year-old male (here after called A02) was admitted to the Second People’s Hospital of Yunnan province, China, with coexisting HBsAg and anti-HBs, identified by clinical examination in December 2015. A02 was followed during July 2016, March 2017, July 2017, and October 2017. The patient provided written informed consent to participate in this study, and the Ethics Committee of the Second People’s Hospital of Yunnan province, according to the Declaration of Helsinki, approved the study protocol. A02 presented HBsAg for at least six months, detectable HBV DNA levels (103 IU/mL), elevated normal serum alanine aminotransferase (ALT) levels (> 40 U/L), and no signs of human immunodeficiency virus, hepatitis virus coinfection or decompensated liver diseases (e.g., variceal bleeding, ascites, or encephalopathy) (Table 1).
| Date | Dec, 2015 | Jul, 2016 | Mar, 2017 | Jul, 2017 | Oct, 2017 |
|---|---|---|---|---|---|
| Treatment | Unt r eatment | ETV + Ganlong capsule 7 months | ETV + Ganlong capsule 15 months | ETV + Ganlong capsule 30 months | ETV + Ganlong capsule 33 months |
| HBsAg | + | + | + | + | + |
| HBsAb | + | + | - | - | - |
| HBeAg | + | - | - | - | - |
| HBeAb | - | + | + | + | + |
| HBcAb | + | + | + | + | + |
| HBV DNA, IU/mL | 7320000 | < 100 | < 100 | < 1000 | < 100 |
| ALT, U/L | 140 | 42 | 41 | 34 | 54 |
| AST, U/L | 70 | 34 | 35 | 31 | 40 |
| TB, umol/L | 19.6 | 12.6 | 16.9 | 24.3 | 14 |
| DB, umol/L | 7.9 | 4.5 | 4.4 | 4.8 | 5.5 |
| IB, umol/L | 11.7 | 8.1 | 12.5 | 19.5 | 8.5 |
3.2. HBV Serology, Liver Biochemistry, and HBV DNA Tests
HBV serological markers were measured using a chemiluminescent micro-particle immunoassay and an Abbott architect immunoassay system (Abbott Laboratories, Abbott Park, IL, US). Liver biochemical and coagulation parameters were determined using an automated chemical analysis system (Beckman Coulter, Fullerton, CA, US). The HBV DNA levels were tested with polymerase chain reaction (PCR) using the CobasAmplicor HBV monitor test (Roche Diagnostics, Mannheim, Germany), with a lower limit of quantification at 100 IU/mL.
3.3. HBV Full-Length Genomic Sequences Acquisition
HBV DNA was extracted from the serum using a Tianlong DNA/RNA virus mini kit (Tianlong, Xian, China). The HBV genome was amplified by PCR in 50 µL of mixture containing 200 ng of template, 2 U of high-fidelity LA Taq DNA polymerase (TaKaRa Bio Inc., Shiga, Japan), 5 µL of 10X LA PCR buffer II (Mg2+ plus), 5 mM of each dNTP, and 10 pmol of each primer, which was designed as described by Gunther et al. (27). The cycling conditions were initial denaturation at 95°C for three minutes, followed by 10 cycles at 94°C for 40 seconds, 60°C for 90 seconds, and 68°C for three minutes, five minutes, and seven minutes successively, with a final extension at 72°C for 10 minutes. The PCR products of 3.2 kb in size were purified using a DNA fragment agarose gel recovery kit (Tsingke, Beijing, China) and cloned in the pGEM-T vector (Promega, Madison, WI, US). The researchers selected one clone per isolate to perform the sequencing. Sixteen positive clones of the sample were sequenced by primers T3 (5’-TAATACGACTCACTATAGGG-3’) and T7 (5’-TTAACCCTCACTAAAGGGA-3’) and four HBV-specific primers P60F (5’-CCTGCTGGTGGCTCCAGTT-3’), P690F (5’-TGTTCAGTGGTTCGTAGGGC-3’), P2300F(5’-AGACCACCAAATGCCCCTATC-3’), and PRES (5’-ATGGGAGGTTGGTCTTCCAAACCTCG-3’), using an ABI 3730 automated sequencer (Applied Biosystems, Foster city, CA, US) (28).
3.4. Sequence Analysis
After assembly and editing using SeqMan Pro software (Bioinformatics Pioneer DNASTAR, Inc., WI, US), full-length genomic sequences were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with default settings. Genetic distances between predefined groups were calculated using Mega6 (http://www.megasoftware.net/, neighbor-joining method, Kimura 2-parameter model, pairwise deletion and 100 bootstrap replicates). Recombination plots were calculated by Grouping Scan analysis with Simmonic 2005 sequence editor V. 1.2 (http://www.virus-evolution.org, 250-bp window size and 50-bp step size, 100 bootstrap replicates). Newly determined genomes were deposited to GenBank (Accession numbers KY470857-KY470872).
3.5. Quasi Species Complexity and Diversity
Shannon entropy (Sn) is used to evaluate quasi species complexity. This was calculated using the formula: Sn = -Si (pilnpi)/lnN; N denotes the total number of clones, and pi denotes the frequency of each clone in the HBV quasi species (29). Quasi species diversity, which is evaluated by the number of synonymous substitutions per synonymous site (dS), the number of non-synonymous substitutions per nonsynonymous site (dN), and the mean genetic distance (d) was calculated, as described elsewhere (29).
3.6. Phylogenetic Tree
First, the researchers collected 35 complete HBV genome sequences from GenBank. Using these data, Mega version 6.0 was employed in phylogenetic reconstruction using GTR + I + G model, which was calculated by ModelTest 3.7 in advance (30).
3.7. Meta-Analysis
A literature search was conducted using PubMed, Embase, and Chinese Biological Medicine databases from inception to January, 2016. The Medical Subject Heading (MeSH) terms “hepatitis B virus”, “HBV”, “pre-S deletion”, “mutation”, “coexistence of HBsAg and HBsAb”, “coexistence of HBsAg and anti-HBs”, and “HBsAg+/anti-HBs+”, “HBsAg+/HBsAb+”, and the individual corresponding free terms served as queries. The studies included in the meta-analysis all met the following criteria: (1) The studies evaluated the relationship between pre-S mutations of HBV and coexistence of HBsAg and HBsAb risk, (2) these were case-control or cohort studies, (3) measurements of HBsAg, anti-HBs, HBeAg, antibodies to hepatitis B e antigen (anti-HBe), and antibodies to hepatitis B c antigen (anti-HBc) were made using standard commercially available methods, and matched patients, who were HBsAg positive yet without anti-HBs served as controls and, (4) Odds ratios with the 95% confidence interval (CI) could be assessed, according to the available data. Data extraction and statistical analysis were performed as described elsewhere (31). Review manager version 5.2 (cochrane collaboration) was used to conduct meta-analyses. Summary odds ratios or relative risks were obtained using a random-effects model. A funnel plot test was performed to examine the publication bias.
3.8. Hot Mutations
HBV strains genotype I HBsAg sequences were collected from GenBank and compared to that derived from patient A02. The study also selected HBV strains from patients, who were HBsAg-positive without anti-HBs for subsequent analysis.
4. Results
4.1. Case Presentation
A 58-year-old Chinese man was admitted to the Second People’s Hospital of Yunnan province in December, 2015 for CHB investigations. He had not received hepatitis B vaccine and had no other known risk factors for viral hepatitis. At the time of admission, virological tests revealed an HBeAg-positive CHB with high levels of HBV-DNA, elevated of ALT (140 U/L), and an unexpectedly low titer of HBsAb (20.2 mIU/mL, with a protective value > 10 mIU/mL). Anti-hepatitis C and D virus antibodies were not detected. After six months of Entecavir (ETV) plus Ganlong capsule treatment, the patient had undetectable HBV DNA, a lower ALT value, and seroconversion of HBeAg (Table 1). However, during the following 26 months, neither a virological response nor a biochemical response was observed, and HBsAb disappeared (Table 1).
4.2. Quasi Species Complexity and Diversity
The general characteristics of the newly determined HBV genomes are summarized in Table 2. The lengths of the complete HBV genomes from the 16 clones ranged from 2 915 bp to 3 125 bp. Quasi species complexity (Sn) and diversity (d, dS, and dN) of 16 clones at the nucleotide and amino acid levels for HBV full-length (at the nucleotide level only), HBsAg, middle HBsAg (MHBsAg), large HBsAg (LHBsAg), polymerase (P), reverse transcriptase (RT), hepatitis B x antigen (HBxAg), and Precore/core (preC/C) are shown in Table 2. A positive correlation between quasi species complexity and gene length was observed, although the HBxAg gene of patient A02 showed the highest Sn. Precore/core showed the highest diversity (d, dS, and dN) among the HBV genes, and HBsAg and MHBsAg showed higher dN than dS values.
| Full-Length | Precore/Core | X | P Value | S | LHbsAg | MHbsAg | RT | |
|---|---|---|---|---|---|---|---|---|
| Quasi species complexity (nucleotide level) | 0.8438 | 0.7443 | 0.8438 | 0.8438 | 0.7264 | 0.8125 | 0.7264 | 0.8438 |
| Quasi species complexity (amino acid level) | 0.6834 | 0.7443 | 0.8438 | 0.6834 | 0.7695 | 0.6834 | 0.8438 | |
| Genetic distance (d) (10-3 substitution/site, nucleotide level) | 15.5487±1.3917 | 22.8399 ± 4.2593 | 15.2814 ± 3.4215 | 12.347 ± 1.3927 | 13.7017 ± 3.0612 | 16.6127 ± 2.7224 | 14.7986 ± 2.9850 | 15.2964 ± 2.6925 |
| Genetic distance (d) (10-3 substitution/site, amino acid level) | 45.1898 ± 10.5444 | 25.2651 ± 7.9837 | 37.3542 ± 5.3460 | 32.5959 ± 7.6600 | 35.5728 ± 5.9240 | 34.5196 ± 7.8691 | 21.5523 ± 5.0000 | |
| dS (10-3 substitution/site) | 28.4329 ± 8.8715 | 24.1701 ± 7.1469 | 22.4781 ± 3.7113 | 11.3313 ± 5.8049 | 18.2784 ± 5.5663 | 13.2368 ± 4.581 | 32.2074 ± 7.2696 | |
| dN (10-3 substitution/site) | 21.1271 ± 4.0186 | 12.1488 ± 3.7788 | 9.0356 ± 1.4089 | 14.4675 ± 3.3567 | 16.031 ± 3.2203 | 15.296 ± 3.4481 | 9.9434 ± 2.4399 |
4.3. Phylogenetic Analyses and Genotyping
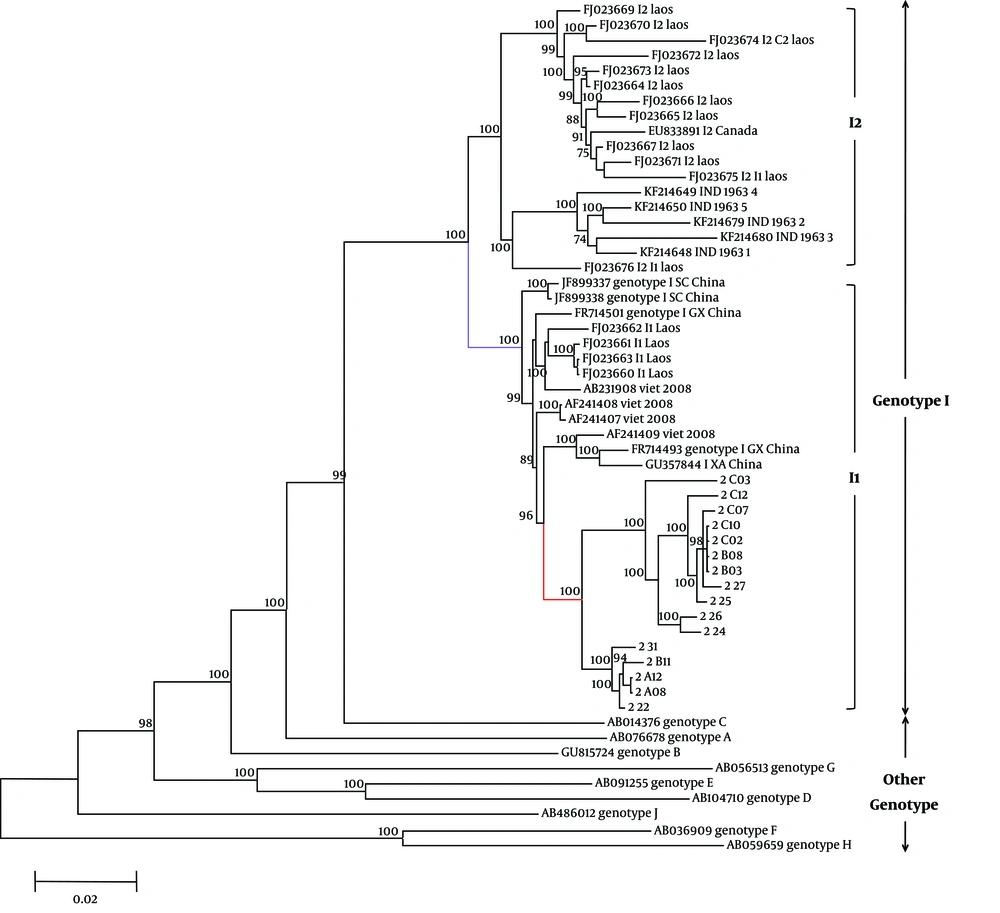
The reconstructed phylogenetic tree showed similar topology to that of a previous study (32), with high bootstrap support (Figure 1). All clones from patient A02 that were divided to two groups were classified within the same sequence clade, with a mean intergroup distance of 2.99% ± 0.17%. This clade clustered with HBV/I strains from China, Vietnam, and Laos (I1), and was separate from that of Indian, Laotian (I2), and other genotype strains. No co-infecting genotypes were identified.
Phylogenetic analysis of 16 full-length HBV genomes from the study patient A02. Sample codes are shown in red. I1 subgenotype is marked in comparative GenBank sequences are designated by the HBV genotype or subgenotype followed by the accession number. The geographic origins of GenBank HBV/I strains showing similarity to the study sequences are also shown (following the accession number). Bootstrap confidence values of 70% are indicated. The ruler shows the branch length for a pairwise distance equal to 0.02.
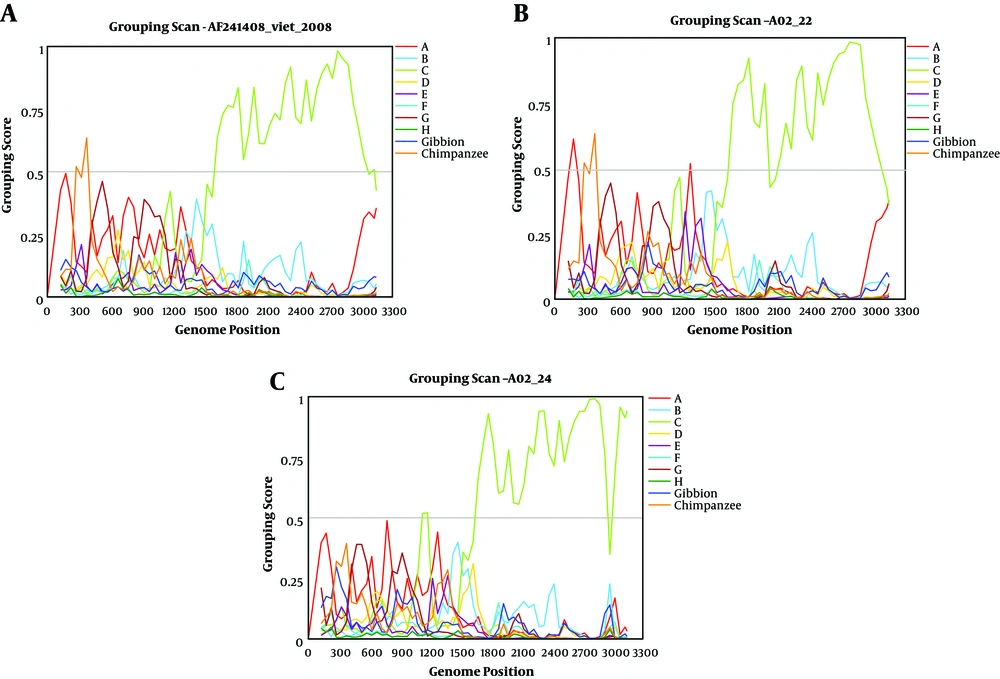
The grouping of A02 with genotype I was confirmed by distance calculation (Table 3). Coincident with intra-genotype distance values, distance values between the A02 and I groups were only 0.044. However, distance values between A02 and other genotypes were > 0.08, which were beyond intra-genotype distance values. All recombination plots were divided to two types, which were in agreement with clones A02 - 24 and A02 - 22 (Figure 2) and coincided with the phylogenetic tree. The plots of A02 - 22 and the Vietnamese strain sequence (AF241408) showed almost the same trend. The genomic region encompassing 1600 bp to 3000 bp was unique to genotype C. However, the first half of the sequence was not consistent with the human or primate HBV genotypes with a low group score (< 0.5), thereby indicating that this is an outgroup. The samples were not clustered with any other reference group. Clone A02 - 24 showed a lower group score (< 0.5) between 2940 bp and 3000 bp, whereas clones A02 - 24 and A02 - 22 had a higher group score (> 0.05) between 1120 bp and 1180 bp (Figure 2).
| A02 (16) | A (74) | B (87) | C (71) | D (53) | E (54) | F (28) | G (37) | H (24) | I (22) | J (1) | Gib (11) | Chi (17) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A02 | 0.015 | ||||||||||||
| A | 0.094 | 0.045 | |||||||||||
| B | 0.102 | 0.106 | 0.052 | ||||||||||
| C | 0.087 | 0.103 | 0.104 | 0.056 | |||||||||
| D | 0.111 | 0.11 | 0.118 | 0.115 | 0.048 | ||||||||
| E | 0.113 | 0.108 | 0.122 | 0.118 | 0.091 | 0.028 | |||||||
| F | 0.147 | 0.154 | 0.154 | 0.152 | 0.156 | 0.154 | 0.056 | ||||||
| G | 0.113 | 0.118 | 0.133 | 0.131 | 0.124 | 0.119 | 0.157 | 0.017 | |||||
| H | 0.147 | 0.15 | 0.154 | 0.149 | 0.151 | 0.153 | 0.089 | 0.154 | 0.013 | ||||
| I | 0.044 | 0.087 | 0.099 | 0.082 | 0.107 | 0.109 | 0.148 | 0.109 | 0.147 | 0.03 | |||
| J | 0.12 | 0.129 | 0.121 | 0.12 | 0.133 | 0.128 | 0.154 | 0.134 | 0.152 | 0.116 | N | ||
| Gib | 0.114 | 0.118 | 0.11 | 0.11 | 0.125 | 0.121 | 0.148 | 0.132 | 0.147 | 0.11 | 0.112 | 0.081 | |
| Chi | 0.11 | 0.109 | 0.117 | 0.114 | 0.112 | 0.099 | 0.145 | 0.115 | 0.141 | 0.105 | 0.118 | 0.103 | 0.055 |
aValues on the diagonal are the mean distances within genotype A - J, chimpanzee genotype (Chi), and gibbon genotype (Gib).
bThe calculation is based on the maximum composite likelihood distance correction method.
4.4. Deletion and Stop Codon Mutations in the HBV Quasi Species
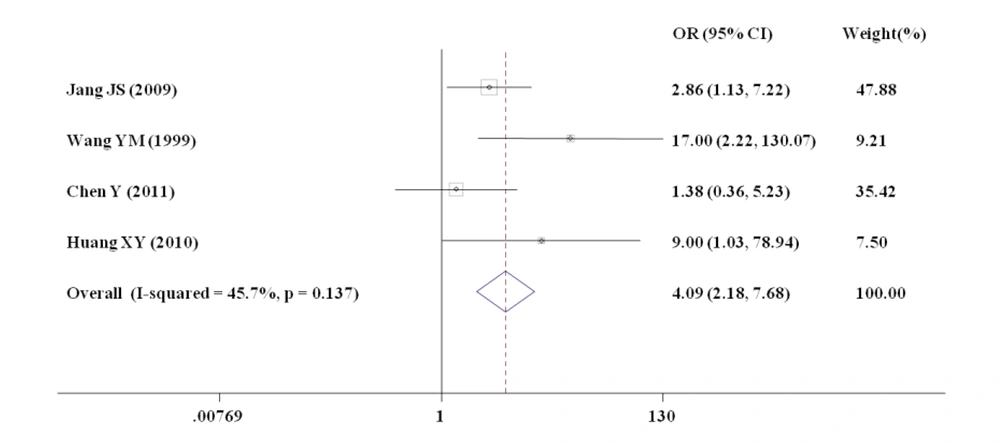
Stop codon mutations were observed in HBsAg and pre-core/core at a mutation rate of 69% (11 of 16 clones, Figure 3). Deletions were only observed in large HBsAg (11 of 16 clones (69%) in PreS1 and 8 of 16 clones (50%) in HBsAg, Figure 3) instead of the three other open reading frames (core, HBxAg, and polymerase). Deletion and stop codon mutations resulted in truncated large HBsAg, HBsAg, and precore/core. This study also assessed the relationship between HBV genome deletions and patients with coexisting HBsAg and anti-HBs by meta-analysis. Overall, 307 papers were selected using the defined criteria. These studies included 508 HBV-infected participants, of which 39 were on the coexistence of HBsAg and anti-HBs. In this meta-analysis, the researchers mainly focused on PreS deletions in this meta-analysis. Finally, four studies evaluating the relationship between PreS deletions mutations in HBV and the risk for the coexistence of HBsAg and anti-HBs were selected. The flowchart of the analysis is shown in Supplementary File Appendix 1. The characteristics of the included studies are shown in Supplementary File Appendix 2. All studies were performed in Asia. Figure 4 indicates a statistically significant association between PreS deletions and the risk for HBsAg and anti-HBs coexistence. The data showed that patients with coexisting HBsAg and anti-HBs had more pre-S HBV deletions than patients without anti-HBs (18.5% versus 9.1%). The overall risk estimate between the coexistence of HBsAg and anti-HBs and PreS deletions was 4.09 (95% CI = 2.18 - 7.68) in a random-effects model. The funnel plot for the studies of PreS mutations indicated no publication bias (Supplementary File Appendix 3).
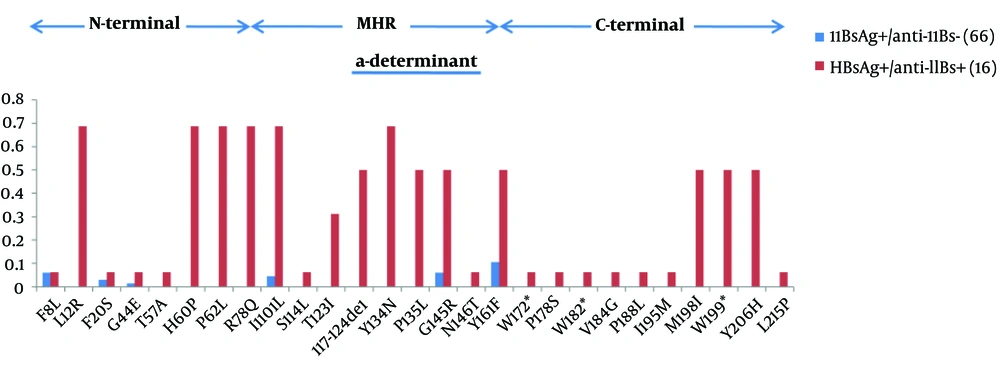
Frequencies of residue substitutions within the S protein. HBsAg is isolated from HBsAg+/anti-HBs+ patients (red bars, n = 16) and solely HBsAg-positive patients (blue bars, n = 66), analyzed in 226 amino acids. Each bar represents the percentage of mutated residues for all clones at each amino acids per group. Residue changes linked to genotypic polymorphisms were not taken into account.
4.5. Assessment of HBV Gene Variability
This study also collected 62 HBsAg genotype I sequences from GenBank, which were from HBsAg-positive patients without anti-HBs. The researchers then conducted HBsAg amino acid substitution analysis (Figure 3). Sequence analyses showed that 68% (11/16 clones) of HBsAg of A02 harbored amino acid substitutions. More than 50% of the clones showed substitutions that were not observed in 62 HBsAg-positive patients without anti-HBs with the following amino-acid changes: 117 - 124del (n = 8), sL12R (n = 11), sH60P (n = 11), sP62L (n = 11), sR78Q (n = 11), sT123I (n = 6), sY134N (n = 11), sP135L (n = 8), sM198I (n = 8), sW199* (n = 8), and sY199H (n = 8) (Figure 3). The most commonly described sG145R substitution was detected in eight clones (50%) from A02, yet only four patients without anti-HBs (6%) had this substitution. These mutations were coupled with previously described deletions. Here, five of the 16 clones showed no deletions and such substitutions. Furthermore, 31 HBx and Pol genotype I sequences from GenBank, which were from HBsAg-positive patients without anti-HBs, were collected. These individuals showed 18 mutations and two deletions of the Pol gene that were not found in HBsAg-positive patients, and three mutations of HBx were only identified in A02. For the Pol gene, most mutations were coupled with deletions.
5. Discussion
In this study, quasi species of the full-length HBV genome of a patient with coexisting HBsAg and anti-HBs were assessed. The threshold for distinguishing different HBV genotypes was set at 8% genetic distance, based on HBV genome sequences (33). As in a previous study (23), the mean genetic distance between A02 and genotype C was 8.2%, while that between A02 and genotype I reached 4.4%, which also showed robust bootstrap support from phylogenetic analysis. The findings suggest that A02 was infected with HBV genotype I, not genotype C (21, 34). Genotype I was proposed to be a recombinant of A/C/G, using the limited SIMPLOT method (5), however, the present study does not supported this theory. The current recombination plot coincides with a reference sequence (AF241408), and the group score of genotype C was higher (> 0.5) in the region of 1500 to 3200 nt, whereas the region 1 to 1500 nt did not provide sufficient evidence to associate the data with any known genotype. Together with genetic distance analysis, it is feasible to designate that patient A02 was infected with HBV genotype I instead of genotype C. Phylogenetic analysis indicated that the 16 clones could be divided to two groups that belonged to subgenotype I1 (Figure 1), which is prevalent in China, Vietnam, and parts of Laos (35). Although increasing evidence for the prevalence of genotype I in Vietnam, Laos, Northeast India, and China (3, 18-21) were found, no reports on liver diseases, such as fibrosis and HCC have been associated with HBV genotype I in these populations. The pathogenic potential and the overall prognosis of this genotype should thus be further evaluated.
Diversity analysis indicated a higher dN than dS in HBsAg and MHBsAg, which may have contributed to the evasion of immunological responses via changes in epitopes sequences. The highest diversity in the pre-core may be associated with its relatively low selective pressure because of the lack of overlap with other genes (36). Interestingly, PreS1 deletions have been reported to be related to the increases in viral replication and disease progression (37). In the current study, 68.75% (11 of 16) of the clones harbored fragment deletions in the PreS1. However, whether these deletions are associated with the coexistence of HBsAg and anti-HBsAg remains unclear. The current meta-analysis suggested that PreS deletions are associated with the coexistence of HBsAg and anti-HBs. This and the current findings indicate the coexistence of HBsAg and anti-HBs may be partially attributable to PreS1 deletions in HBV quasi species. Because many antigenic epitopes are identified on PreS1 (38), deletions involving PreS1 may have resulted in decreased immunogenicity in patient A02.
In the majority of cases, the HBV immune escape variants were found in the “a” determinant of the HBsAg spanning amino acids (aa) 124 to 147, particularly the arginine to glycine substitution at aa position 145 (39). Furthermore, HBV quasi species from patient A02 had significantly higher aa variability than controls (MHR antigenic loops and C-terminal regions). These mutations were mostly located within the cysteine-rich “a” determinant of the HBsAg MHR, which is considered as the main target of humoral responses (25). Moreover, a 117 to 124 aa deletion was detected in patient A02, which was situated just on the first loop of the “a” determinant, thereby possibly inducing modifications (40). Mutations involving highly conserved amino acids within the HBsAg of patient A02 were detected. Interestingly, most of these mutations were coupled with deletions in PreS1. It is possible that PreS1 deletions might have changed the antigenicity of some clones, thus, allowing them to evade humoral responses, which in turn causes HBV quasi species to undergo relaxed selective pressure, which is followed by an accumulation of amino acid substitutions in HBsAg. It is not clear whether deletions in preS/S decrease the fitness of HBV quasi species. A relative molar ratio of LHBsAg to HBsAg is essential for the release of replicating viral particles and to the control the cccDNA formation rates (41). The balanced expression of envelope proteins from five strains encoding wild-type proteins appears to explain why quasi species harboring several deletions in preS/S were present in patient A02. Although the patient experienced HBeAg seroconversion, the loss of HBV DNA, and subsidence of the hepatic inflammatory activity, because of the pronounced diversity of the HBV genome quasi species (especially in preS/S), there was also a loss of anti-HBs and HBsAg that lasted through a large part of the later stage of treatment.
This is the first report on an HBV quasi species population with genotype I from a male patient with coexisting HBsAg and anti-HBs infected. Among all HBV genes, procore harbored the highest diversity in the A02 quasi species. The A02 quasi species showed a propensity for PreS1 deletions, which coincides with the results of the current meta-analysis, together with the complexity of HBsAg variants, are related to the coexistence of HBsAg and anti-HBs. In summary, the presence of these exceptional variants in HBV isolates from a patient infected with HBV genotype I could explain the coexistence of HBsAg and anti-HBs and the resistance to current antiviral therapies.