1. Background
Non-alcoholic fatty liver disease (NAFLD) is considered one of the major causes of chronic liver disease characterized by the accumulation of triglycerides and other fats in hepatocytes of the liver parenchyma without a history of excessive use of alcohol, infection by viruses, hereditary disorders, or the use of steatogenic drugs (1). NAFLD includes a wide spectrum of histological and clinical changes ranging from hepatic steatosis (fatty liver) to non-alcoholic steatohepatitis (NASH) featured by hepatocellular injury and inflammation, which can often lead to fibrosis and liver cirrhosis or hepatocellular carcinoma (HCC) (2, 3). A number of studies have reported that more than one billion people and some others have estimated that 25% of the adult populations in the world have NAFLD (4, 5). Obesity, type 2 diabetes mellitus (T2DM), and dyslipidemia are considered the major causes of NAFLD. Therefore, the prevalence of NAFLD is expected to increase due to a number of components of metabolic syndrome (6, 7). Furthermore, a range of risk factors such as high-calorie diet, eating habits, inactive lifestyle, genetic susceptibility, and racial and ethnic background can also contribute to the increased prevalence of NAFLD (8-11).
Recent studies have reported that mitochondrial dysfunction and oxidative stress are the main players in the pathogenesis of NAFLD and its progression (12).
Mitochondria are known as the main site of oxidative phosphorylation and fatty acid ß-oxidation and are also involved in the formation of reactive oxygen species (ROS) in living cells (13, 14). The mutation rate is higher in mtDNA than in nuclear DNA (nDNA) in mammals due to the lack of protective histones and DNA repair mechanisms, as well as the close vicinity to the ROS production site (15). Mutations in different parts of mtDNA such as D-loop may affect mitochondrial functions and increase the risk of susceptibility to different diseases such as cancer and metabolic and neurodegenerative disorders (16, 17). Therefore, it is hypothesized that the deterioration of mitochondrial function and disruption of fatty acid beta-oxidation will lead to the significant accumulation of free fatty acids and other lipid molecules in the cytosol. Therefore, excessive hepatic lipid accumulation in NAFLD patients can affect the mitochondrial oxidation, which leads to oxidative damage to mitochondrial proteins, DNA, and other cellular components, and can finally cause cell death and progression of the disease to NASH (12, 18, 19). According to the above-mentioned explanations, it has been suggested that due to the role of hepatic mitochondria in the development and pathogenesis of NAFLD, the disease might be considered a mitochondrial disease (20).
Currently, the use of mtDNA mutations and/or polymorphism patterns may serve as a biomarker (15) and until now, more than several hundred identified mutations in the Mitomap database have been reported in a wide variety of human disorders (21).
2. Objectives
To date, a few studies have focused on the relationship between mtDNA and NAFLD.
This study evaluated 4977-bp deletion and the D-loop region variation for the first time in Iranian patients with NAFLD with the aim of evaluating the possible relation of this common deletion and D-loop region mutations with NAFLD.
In order to determine whether a correlation exists between NAFLD and the amount of deleted 4977-bp and evaluate whether this deletion can serve as an ideal biomarker for early detection, we examined 43 human blood samples in NAFLD patients to detect the mtDNA 4977-bp deletion.
Since D-loop is the most polymorphic region in mitochondria and vulnerable to oxidative damage due to ROS formation, it can be hypothesized that specific SNPs in this region may act as an important component in the pathogenesis or progression of NAFLD. Therefore, to investigate the possible role of SNPs in NAFLD predisposition, we analyzed the D-loop sequence of 43 Iranian NAFLD patients and 156 normal controls for the possible presence of variations.
3. Methods
3.1. Patients and Samples Population
The study population consisted of 43 NAFLD patients with a mean age of 42.37 ± 9.66 years (Table 1) undergoing laparoscopic sleeve gastrectomy and referring to Khatam-al-Anbia Hospital, Tehran, Iran, during the period from May 2014 to November 2015. Blood samples were collected from NAFLD diagnosed subjects based on imaging technique and histological evidence. Subjects who met any of the following criteria were excluded from the study: A secondary cause of chronic liver disease including alcohol intake, infection with hepatitis B and C viruses, Wilson’s disease, taking medications such as amiodarone, methotrexate, tamoxifen, and corticosteroids, a family history of NAFLD, and diabetes. The control group consisted of 156 Iranian healthy subjects whose clinical, laboratory, and imaging findings were completely normal. This case-control study was ratified by the Scientific and Ethics Committee of Hamadan University of Medical Sciences. All of the participants were informed of the objectives of the study and they signed written informed consent forms.
3.2. Anthropometric and Biochemical Measurements
Body mass index (BMI) was calculated as body weight (kilograms) divided by height squared (meters) (BMI = weight/height2). Blood pressure (BP) was measured two times in the seated position by a trained nurse using desktop mercury column sphygmomanometer, and then the average of the two BP measurements was used as the final BP. Aspartate transaminase (AST), Alanine transaminase (ALT), triglyceride, cholesterol, HDL-cholesterol, and fasting blood glucose levels were determined by standard enzymatic methods using auto-analyzer. In addition, the Friedewald formula was used to determine the LDL-cholesterol levels.
3.3. DNA Extraction
Peripheral blood samples were taken from the subjects and DNA was extracted after the lysis of white blood cells using a Macherey-Nagel NucleoSpin® Blood DNA extraction kit, as per manufacturer’s instructions. Then, the extracted genomic DNA was kept at -20°C. After completing DNA extraction from blood samples, the qualification of DNA samples was done by a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and then, the isolated DNA was kept at -20°C.
3.4. Detection of mtDNA 4977-bp Deletion
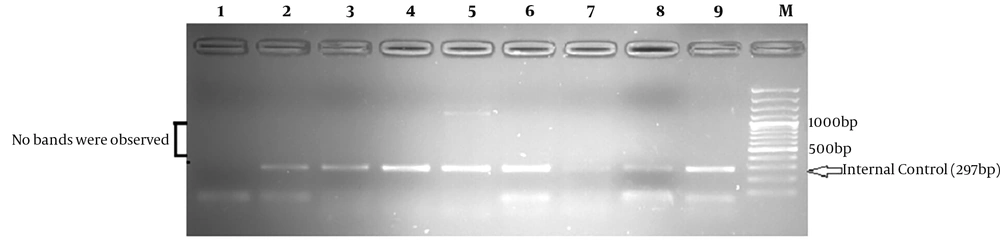
Multiplex PCR was performed for the detection of 4977-bp deletion in DNA samples with the use of the following primer sets: ONP86/ONP89 and ONP25/ONP74 (22). It should be noted that Primer-Blast program (http://www.ncbi.nlm.nih.gov/tools/primer-blast) was used to check primer specificity. The PCR cycling conditions to detect this deletion consisted of initial denaturation at 94°C for three minutes, followed by 35 cycles at 95°C for 40 seconds, annealing at 57.5°C for 40 seconds, extension at 72°C for 40 seconds, and a final extension at 72°C for seven minutes. As an internal control, a normal mtDNA fragment (297 bp) was amplified in all samples by the use of the first set of primers (86/89) for the PCR analysis while the second set (25/74) was used to anneal outside of the deletion. The presence of a 497-bp fragment on a 1.5% agarose gel confirmed the expression of 4977-bp deletion. Wild-type mtDNA (mtDNA with no large deletion) would not reveal this fragment on the agarose gel.
3.5. PCR-Sequencing Analysis
According to the following PCR protocol, ONP 98/79 primers were used for the amplification of mtDNA D-loop region (15): Pre-denaturation at 95°C for 3 minutes, followed by 35 cycles of 95°C for 40 seconds, 57.5°C for 40 seconds, and 72°C for 40 seconds, and a final extension step of 72°C for 7 minutes. After electrophoresis, the amplification of the D-loop region was confirmed by the appearance of a single fragment 1558-bp on a 1% agarose gel. Then, each PCR product was sequenced with the use of a sequencer (gene Fanavaran, Macrogene Seoul, Korea). DNA sequences qualification was assessed by Finch TV version 1.4.0 (Geospiza Inc.). Finally, the findings were compared with a human mitochondrial database (http://www.mitomap.org/) (23).
3.6. Statistical Analysis
SPSS version 18 statistical software was used for all statistical analyses in this study. Means ± standard deviation (SD) or median with interquartile range (IQR) were used to express the quantitative data. The t-test and one-way ANOVA as parametric tests and Wilcoxon-Mann-Whitney test as a nonparametric statistical test were used for the comparison of normally and non-normally distributed data between the groups, respectively. Logistic regression analysis was conducted to adjust for confounders. Pearson correlation was used to investigate the relationship between the number of mtDNA D-loop mutations and the study variables. P values of < 0.05 in all tests were considered statistically significant.
4. Results
According to the results, patients with NAFLD had significantly higher values of BMI (P < 0.05) but no significant difference was found between these two groups in age and gender (P > 0.05).
4.1. Mitochondrial Common Deletion
With the use of the multiplex PCR, the analysis of D-loop region confirmed that the 497 bp product, which corresponds to 4977-bp mtDNA deletion, was not detected in any of the collected blood samples from NAFLD patients and controls (Figure 1).
4.2. Mitochondrial D-Loop Polymorphisms and Correlation with Patient’s Age, Gender, and BMI
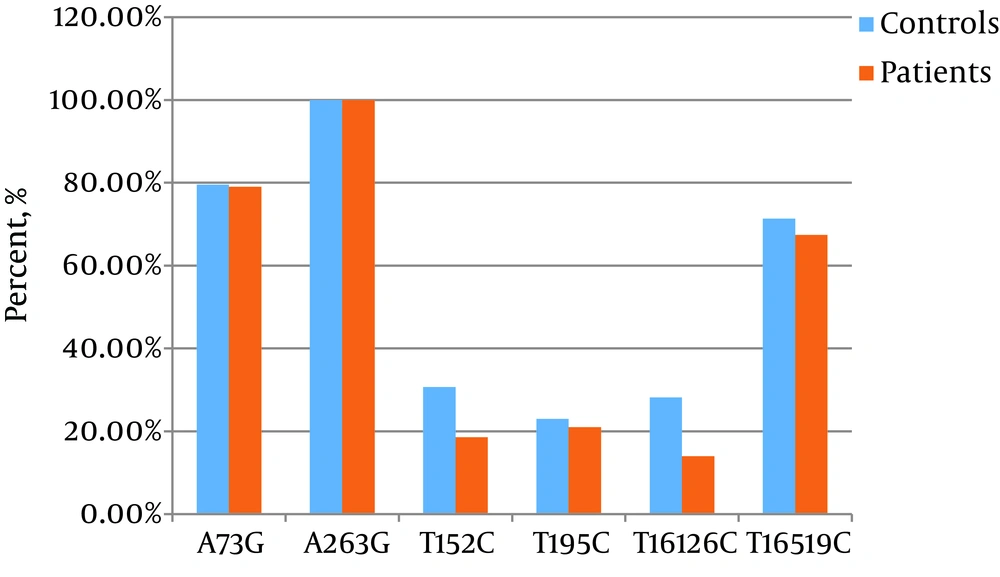
The mtDNA D-loop region was assessed by sequencing for the comparison of D-loop variants between the two groups, which resulted in 94 different variations including two deletions, four insertions, and 88 single nucleotide polymorphisms (SNPs). Among the detected variations, 16 variations including T119C, C258T, T279C, T334C, A335G, C431A, T477C, T16324C, C16344T, A16078G, C16082T, A16220C, T16263C, 16221 ins C, 16171 ins T, and 523 (CA)3 ins were only found in NAFLD patients. The most common variants, A73G, A263G, T195C, T16126C, T152C, and T16519C, were detected in both healthy controls and NAFLD patients (Figure 2). These polymorphisms were not significantly correlated with NAFLD (P < 0.05).
There was a statistically significant difference between NAFLD patients and controls in six variations (P < 0.05; T334C, C16111T, A16220C, C16266T, c16221ins, and A248del) (Table 2). These results remained statistically significant after adjusting for age, gender, and BMI.
| Mutation | Region | Controls, No. (%) | NAFLD Patients, No. (%) | P Valueb | Odds Ratio (95% CI)c |
|---|---|---|---|---|---|
| A248del | HV2 | 2 (1. 28) | 3 (6. 97) | 0.048 | 5.34 (1.01 - 28.07) |
| T334C | HV2 | 0 (0) | 3 (6. 97) | 0.030 | 27.07 (1.36 - 534.2) |
| C16111T | HV1 | 1 (0. 64) | 3 (6. 97) | 0.027 | 8.95 (1.28 - 62.57) |
| A16220C | HV1 | 0 (0) | 3 (6. 97) | 0.030 | 27.07 (1.36 - 534.2) |
| C16221ins | HV1 | 0 (0) | 4 (9. 30) | 0.017 | 35.36 (1.88 - 676.2) |
| C16266T | HV1 | 1 (0. 64) | 3 (6. 97) | 0.027 | 8.95 (1.28 - 62.57) |
List of Single Nucleotide Polymorphism in Healthy Controls and NAFLD Patientsa
D-loop sequencing also showed that among the observed variants in the NAFLD group, 16171 ins T and 16221 ins C were novel. Based on the results, C16266T SNP was more frequent in NAFLD patients than in the controls. In addition, our results indicated that all D-loop variations in NAFLD patients were homoplasmic.
Our results also showed that there was no statistically significant difference in mutations discussed above between women and men (P = 0.501), as well as age groups (P = 0.125). In addition, no significant relationship was found between these mutations and other parameters such as BMI after adjusting for age and gender (P > 0.05).
The analysis of D310 region in mtDNA showed three patterns of C variation (C7TC6, C8TC6, and C9TC6), but no significant difference (P > 0.05) was reported in this variation between NAFLD patients and controls (Table 3).
The frequency of the CA-repeats including CA4, CA5, CA6, and CA8 was 16.2%, 79%, 2.3%, and 2.3% in NAFLD patients, respectively. Among these CA-repeats, CA8 was identified only in one NAFLD patient. However, in the comparison of the two groups, no significant difference was found in the distribution pattern of 514 523 (CA) n (P > 0.05) (Table 4).
| Group | 514 523 (Ca) n | |||
|---|---|---|---|---|
| CA4 (%) | CA5 (%) | CA6 (%) | CA7 (%) | |
| Controls, n = 156 | 34 (21.79) | 116 (74.35) | 6 (3.84) | 0 |
| NAFLD patients, n = 43 | 7 (16.27) | 34 (79.06) | 1 (2.32) | 1 (2.32) |
| P value | 0.428 | 0.526 | > 0.999 | 0.216 |
Distribution Patterns of 514 523 (CA) n Variation in NAFLD Patients and Controls
5. Discussion
Although the definite pathogenesis of NAFLD in the lack of an exact etiology remains challenging, experimental evidence indicates that defects in mtDNA due to oxidative stress may have an important role in the NAFLD pathogenesis. mtDNA is constantly subjected to oxidative damage due to ROS production. On the other hand, increasing oxidative stress may be a secondary factor that results in mitochondrial dysfunction and therefore, mtDNA is more prone to mutations than nuclear DNA (nDNA) (24). Mutations of mtDNA might cause various human mitochondrial disorders. Among the mutations in mtDNA, 4977 bp deletion is the most prevalent large deletion, which was not studied in NAFLD patients before. Our results showed that this deletion was not detected in any of the extracted DNA samples from peripheral blood. Indeed, several studies have reported that the role of point mutations is more important than that of large mtDNA deletions in mitochondrial diseases. In agreement with our results, some previous studies have shown that 4977-bp deletion was not observed in blood samples of patients (25, 26). However, some other findings are in contrast to the results of our study (22). The 4977-bp deletion was observed in 18.6% (8/43) of the liver tissue samples obtained from patients with NAFLD (27). It is also important to consider that the frequency of common large deletion is different in human tissues and the major reason for the formation of these mtDNA deletions in various tissues is still not clear (28).
D-loop as a controlling region for mtDNA transcription and replication is the most variable part due to the high mutation rate; it is also more susceptible to numerous damages. A variation in this region might affect the initiation of mtDNA replication and can also induce ROS overproduction due to alterations in the mitochondrial respiratory chain function (29, 30).
In the present study, multiple point mutations were observed in the mtDNA D-loop region. The analysis of this regulatory region revealed 94 variations, 80 of which were seen in both groups and 16 variations occurred only in NAFLD patients. Our results showed that SNPs including A263G, A73G, T195C, T152C, T16126C, and T16519C were the most common variants in both groups but the difference in terms of these mutations was not statistically significant between NAFLD patients and the control group. The A73G variant has been detected in hearing loss, Alzheimer’s disease, and hypertrophic cardiomyopathy. The A263G variant has been reported in hypertrophic cardiomyopathy, idiopathic cardiopathy, and muscle pathology (31). The T195C variant has been observed in repeated pregnancy loss and psychiatric disorders (21). The T152C variant has been associated with an increased risk of respiratory morbidity in children (32). Moreover, this variant is associated with the presence of a fusion gene (promyelocytic leukemia retinoic acid receptor α (PML RARα) in patients with acute myeloid leukemia (AML) (33). The C16519T and T16126C variants have been correlated with an increased risk of Huntington disease (HD) (34) and the T16519C variant has been also reported as being associated with a predisposing genetic factor for diabetes mellitus and gastrointestinal disorders due to its ability to worsen the pancreatic cancer prognosis (35). The T119C variant affects the conserved regions involved in the mtDNA replication process (36). The T477C mutation was found in the brain tissues of Alzheimer’s disease patients (37). The T16263C variant has been reported in Hepatocellular carcinoma (38).
A335G and C16344T have been observed in patients with repeated pregnancy loss and endometriosis, respectively, but these two mutations were not statistically significant in the diseases (21, 39). Other significant mutations have not been previously reported in NAFLD patients in similar studies. Apart from these, in contrast to other variations in D-loop, six SNPs including T334C, C16111T, A16220C, C16266T, 16221Cins, and A248del indicated significant correlations with NAFLD and their frequency was higher in patients with NAFLD than in the control group (P < 0.05). No positive association has been previously reported between these variants and NAFLD and further studies are needed to confirm this association. We detected two novel variants (16171 ins T and 16221 ins C) in NAFLD patients.
In this study, we also observed that C →T and T → C transitions were the most distributed mutations in D-loop. This type of mutation is associated with aging (40) or carcinogenesis in a variety of organs (41). In concordance with our results, Kavahara et al. found that mtDNA mutations in NASH patients were of transition type; they might also play an important role in the pathogenesis of the disease and cause to mitochondrial disorders, as well. As far as we know, there has not been reported any evidence for the relationship between these variations and NAFLD. The role of these mitochondrial variants in NAFLD is still unknown and further studies are needed to verify this hypothesis.
Mitochondrial microsatellite instability has been reported in many different types of cancer. D310, as the most common mtMSI within the D loop region, is a poly C repeat stretch and the numbers of this repeat vary from person to person. Recently, this region, which has a key role in the formation of persistent RNA-DNA hybrid, has been recognized in some disorders (42) but there was no statistically significant difference between NAFLD and controls in the prevalence of D310 mutations in our study.
The (CA) n dinucleotide repeat polymorphism is one of the most mtDNA variations located in mt514 mt523 (43, 44), which is a powerful tool in forensic identification (45). Our results showed no significant difference between the two study groups and no relationship between the (CA) n polymorphism in the D-loop region and NAFLD. In agreement with our results in this study, Ye et al. also did not report any relationship between the mentioned polymorphism and the risk of breast cancer (46).
In conclusion, this is the first case-control study on evaluating the mtDNA D-loop region in a sample of Iranian population with NAFLD. We detected two novel mutations in NAFLD patients. A statistically significant association was found in six SNPs between patients and controls. No significant difference was found between the two groups in D310 mutations. The 4977-bp deletion was not found in any of the DNA samples extracted from peripheral blood of 43 NAFLD patients. Transitions C→T and T → C were the most common types of mutations in the D-loop region. As some mutations in D-loop are frequently seen in NAFLD patients, it seems that mitochondrial variants by affecting the mitochondria may have a secondary role in the pathogenesis or progression of NAFLD. However, further clinical trials with appropriate sample size and longer follow-up periods are required to verify the findings of the present study.