1. Context
Hepatocellular carcinoma (HCC), which is the most common primary liver cancer, ranks the sixth among all cancers and is the second leading cause of cancer death with a high mortality ratio (1). In 2012, there were approximately 782,000 newly diagnosed cases and 746,000 deaths worldwide, and the quantity is projected to increase in the future (2). The development of HCC is closely related to the presence of chronic liver diseases. Alcohol, HBV, and hepatitis C virus (HCV) significantly increase the risk of HCC (3, 4). Among the three mentioned major risk factors, it has been reported more than 50% of HCC cases worldwide are related to chronic HBV infection (5, 6). At present, the mechanism of HBV-associated HCC is still unclear and thought to be a multi-factorial process including both direct and indirect effects, such as the integration of HBV DNA into the host genome, the increased level of oxidative stress, the regulatory proteins HBx on cytoplasmic signaling pathways, and so on (7). However, developing HCC is still an event of much little probability for patients with HBV. It has been reported that annual HCC incidence is only 0.3% - 0.6% in non-cirrhotic patients, while only 2.2% - 3.7% of compensated cirrhotic patients finally develop HCC among untreated patients, in which Asian patients dominate (8). Thus, there must be some other mechanisms to affect the process. Besides extrinsic factors like hepatitis delta virus (HDV) (9), alcohol (10), and aflatoxin (11), some evidence has demonstrated that there are some genetic factors contributing to HCC susceptibility (12). Through a genome-wide association study (GWAS), Gu et al. (13) found that the polymorphism of CTLA-4 gene might increase susceptibility to hepatitis B-related HCC. Chou et al. (14) reported the relationship between hepatitis B virus enhancer II/basal core promoter sequence variation and the risk of HCC. Jiang et al. (15) pointed out that genetic variants in STAT4 and HLA-DQ genes conferred the risk of HBV-related HCC.
Recently, a study revealed that the serum level of 25-hydroxyvitamin D (25(OH)D) is inversely associated with the risk of HCC (16). Although there is insufficient epidemiologic research to explore the relationship between vitamin D and HCC, much experimental evidence in vivo and in vitro revealed that vitamin D and its analogs inhibited the growth of HCC (17, 18). In a previous clinical trial, EB 1089 (an analog of vitamin D) was applied in 56 patients with inoperable HCC. Though no controls were included in the study to give a convincing answer, tumors were shrunk in two patients and other 12 patients were stable (19). Among all vitamin D metabolites, 1α,25-(OH)2D3 is the most active form. Through its binding to vitamin D receptor (VDR), the expression of corresponding genes can be modulated (20), which leads to antiproliferation, anti-inflammatory response, pro-differentiation, pro-apoptosis, and immune regulation in specific cells and tissues (21, 22).
The VDR gene is reported to be located on chromosome 12q12-q14 while the Fok I polymorphism site is located on the 5’ end of the VDR gene (23). Most studies have focused on four single nucleotide polymorphisms (SNPs) of VDR: TaqI (rs731236), Fok I (rs2225870), ApaI (rs7975232), and BsmI (rs1544410). Among the four sites, only can Fok I polymorphism influence the VDR protein structure by changing off the transcription initiation site (24). Due to the transition of a single nucleotide from T to C in exon 2 at the 5’-end of the VDR gene, protein translation can start from the first initiation codon ‘f’ rather than from the second codon ‘F’. The variant VDR protein might have a less effective function, which was hypothesized to be related to the increased susceptibility to cancer or to a more aggressive disease (23).
Since VDR plays an important role in the action of vitamin D and the possible relationship between vitamin D and hepatocellular carcinoma, it is not surprising to find some interest attracted to SNPs of the VDR gene and HBV-related HCC risk (25). Through the consideration and analysis of the relationship between HCC and VDR polymorphism, first, we can distinguish higher risk groups from a previous population at risk and thus, can revise a more effective screening program. Besides, the discovery of additional genetic risk factors will draw much attention to the role of vitamin D in the generation of HCC, which can aid in a more complete understanding of the interaction between HBV and HCC. What’s more, the differentiation of VDR may give a potential target for new drugs. Among all the SNPs, Fok I polymorphism (rs225870) is more widely involved. Most researchers support the association between Fok I polymorphism and the risk of HBV-related HCC (25-27), but the conclusion of each researcher may be limited due to the low power of individual studies. Therefore, it is significant for us to conduct this meta-analysis to achieve a more accurate conclusion.
2. Methods
2.1. Data Sources and Search
We searched PubMed/Medline, EMBASE, Cochrane library, VIP Database for Chinese Technical Periodicals, WanFang Data, Sino Med, and Chinese National Knowledge Infrastructure database using keywords “(VDR OR vitamin D receptor)” AND “(hepatocellular carcinoma OR HCC OR liver cell carcinoma OR adult liver cancer OR hepatoma)” AND “(polymorphism OR mutation OR variant)”. The search was carried out in July 2018. Publications were limited to those reporting results from human studies. All abstracts were checked independently by two investigators and the reference list of the relevant publications was manually searched.
2.2. Study Selection: Inclusion and Exclusion Criteria
Studies reporting the relationship between VDR polymorphism and HBV-related HCC risk were included. Exclusion criteria were: (1) Studies lacking data of the association of Fok I polymorphism with HCC risk, (2) studies lacking a case group of HBV-related HCC patients, (3) studies lacking a control group of HBV-infected patients, and (4) duplicated studies from the same institution and published at nearly the same time.
2.3. Quality Assessment and Data Extraction
The methodological qualities of the selected studies were assessed through the Newcastle-Ottawa scale (NOS) for observational studies. Every study meeting the criteria was carefully checked by two investigators independently for gathering the following information: First author, publication year, race, country, the source of controls, sample size, and value of Hardy-Weinberg equilibrium (HWE). If any disagreement existed, a third investigator was involved to reach consensus on all of the items.
2.4. Data Synthesis and Analysis
χ2 and Ι2 statistical tests were used to evaluate the heterogeneity between studies. When the Ι2 > 50% and P < 0.05, the heterogeneity is significant and a random effects model is recommended. Otherwise, a fixed effects model is used in homogeneous studies.
Pooled odds ratio (ORs) were used to evaluate the strength of the relationship between the Fok I polymorphism and HCC risks, and the results were reported with 95% confidence intervals (CI). If a P value < 0.05, it was indicated the significant difference of ORs through a Z-test. To get a more detailed answer, all the six genetic models were used in our study: Codominant model (OR 1) (ff vs. FF), codominant model (OR 2) (Ff vs. FF), dominant model (ff vs. FF/Ff), recessive model (ff/Ff vs. FF), codominant model (OR 3) (ff vs. Ff), and allele genetic model (f vs. F). All statistical analyses were performed using STATA package version 12.0.
3. Results
3.1. Search Results and Included Studies
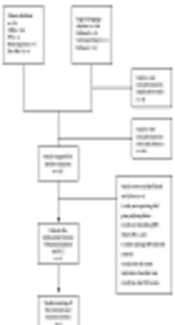
A total number of 198 articles were identified in the electronic search in foreign databases such as PubMed/Medline, EMBASE, and Cochrane Library, and Chinese databases such as CNKI, VIP, Wanfang Data, and SINOMED (Figure 1). Three studies from 2013 to 2016 were finally included in our analysis (25-27). The numbers of HCC cases and controls were 728 and 920, respectively (Table 1). The HWE was meet in the distribution frequency of all the genotypes in control groups, which indicated that all samples were from the same Mendelian population and the sampling bias was low. When being assessed using the NOS, all studies gained a score of ≥ 5 stars, with the highest score of six stars (Table 2).
Abbreviations: HB, hospital-based; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HWE, Hardy-Weinberg equilibrium; NOS, Newcastle-Ottawa scale; Y, yes.
| Newcastle-Ottawa Scale | Studies | ||
|---|---|---|---|
| Yao et al. (27) | Peng et al. (26) | Mohammed et al. (25) | |
| Selection | |||
| Is the case definition adequate? (maximum: *) | * | * | * |
| Representativeness of the cases (maximum: *) | * | * | * |
| Selection of controls (maximum: *) | - | - | - |
| Definition of controls (maximum: *) | * | * | * |
| Comparability | |||
| Comparability of cases and controls on the basis of the design or analysis (maximum: *) | * | - | * |
| Exposure | |||
| Ascertainment of exposure (maximum: *) | - | - | - |
| The same method of ascertainment for cases and controls (maximum: *) | * | * | * |
| Non-response rate (maximum: *) | - | ||
| Total score | ***** | **** | ***** |
3.2. Pooled Results
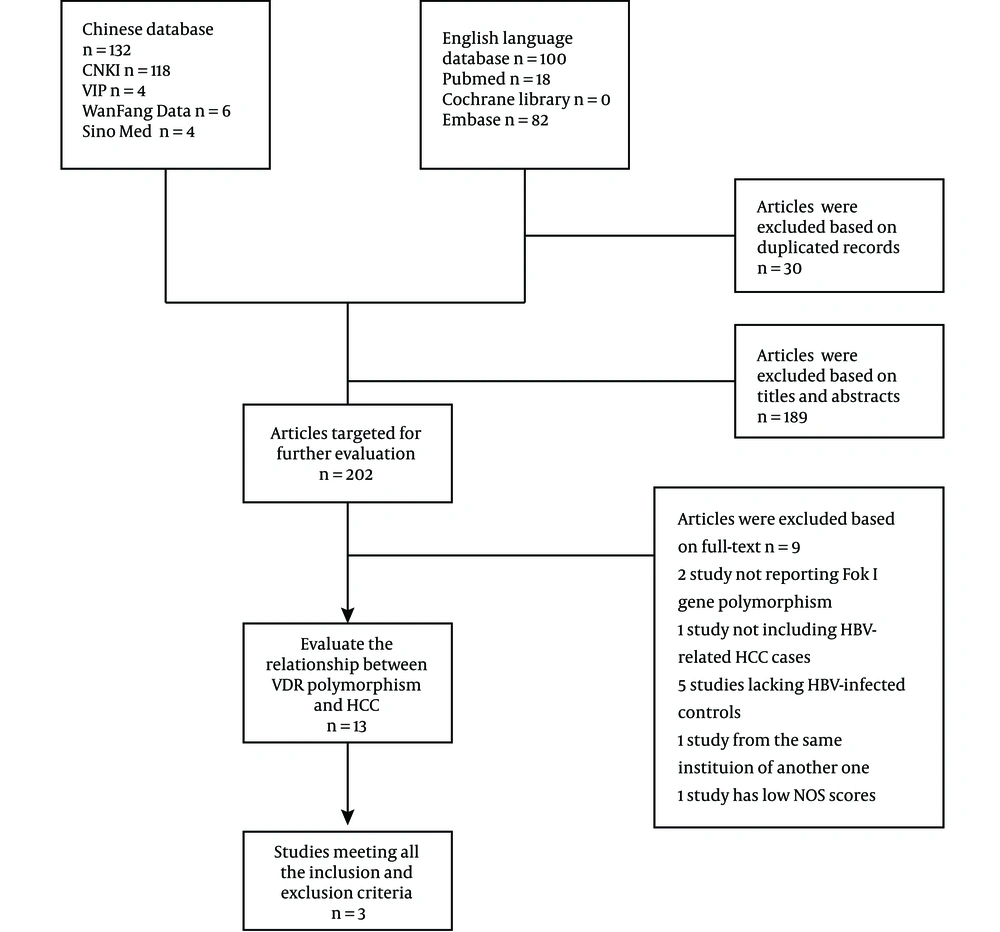
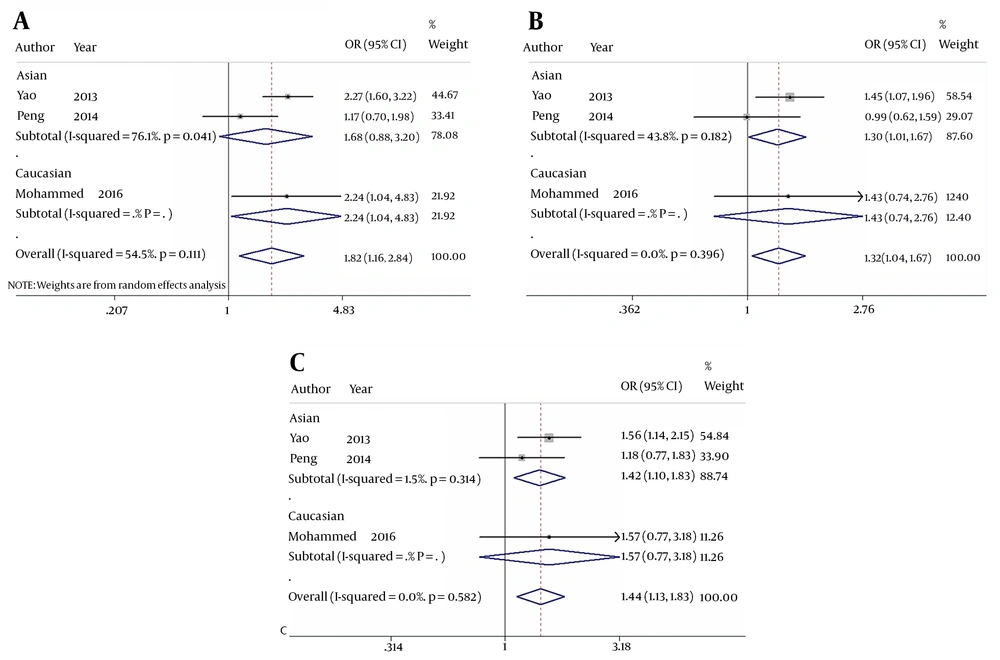
When comparing HBV-related HCC cases with HBV-infected controls, our data showed that five genotypes of Fok I polymorphism significantly increased the risk of HCC in the overall population carrying f allele: ff vs. FF: OR = 1.816, 95% CI = 1.161 - 2.841, P = 0.009 (Figure 2A); Ff vs. FF: OR = 1.315, 95% CI = 1.037 - 1.667, P = 0.024 (Figure 2B); ff/Ff vs. FF: OR = 1.504, 95% CI = 1.206 - 1.876, P < 0.001 (Figure 3A); ff vs. FF/Ff: OR = 1.591, 95% CI = 1.270 - 1.992, P < 0.001 (Figure 3B); f vs. F: OR = 1.375, 95% CI = 1.075 - 1.759, P = 0.011 (Figure 3C); and ff vs. Ff: OR = 1.435, 95% CI = 1.127 - 1.827, P = 0.003 (Figure 2C). Since OR1 > OR2 > 1 and OR1 > OR3 > 1 (OR1 = ff vs. FF; OR2 = fF vs. FF; OR3 = ff vs. Ff), then a codominant model was suggested in Fok I polymorphism28. These results suggest that Fok I genotypes fF and ff and allele f show a close relationship with an increased risk of HBV-related HCC.
The forest plot of the association between Fok I polymorphism and HBV infection. A, B, and C represent the codominant model (OR 1) (ff vs. FF), codominant model (OR 2) (Ff vs. FF), and codominant model (OR 3) (ff vs. Ff), respectively. Odds ratios and respective 95% confidence intervals for the different studies included are shown. The heterogeneity was checked by the chi-square-based Q test.
The forest plot of the association between Fok I polymorphism and HBV infection. A, B, and C represent the recessive model (ff/Ff vs. FF), dominant model (ff vs. FF/Ff), and allele genetic model (f vs. F) respectively. Odds ratios and respective 95% confidence intervals for the different studies included are shown. The heterogeneity was checked by the chi-square-based Q test.
3.3. Heterogeneity
When comparing HBV-related HCC cases with HBV-infected controls, a marginal heterogeneity was found between studies in the codominant model (OR1, f vs. F:I2 = 60.9%, P = 0.078) and allele contrast (ff vs. FF:I2 = 54.5%, P = 0.111). Thus, we had to choose a random effects model to combine the values from the included studies. For the rest models that showed no obvious heterogeneity (Ff vs. FF:I2 = 0.0%, P = 0.396; ff/Ff vs. FF:I2 = 39.2%, P = 0.193; ff vs. FF/Ff:I2 = 31.0%, P = 0.235; and ff vs. Ff:I2 = 0.0%, P = 0.582), a fixed effects model was used in these studies and the one with the same situation.
4. Conclusions
It is well known that alcohol, HBV, and HCV are the main risk factors of HCC (3, 4). However, the underlying causes remain unclear. Apart from extrinsic factors like HDV (9), alcohol (10), and aflatoxin (11), there is increasing research on the contribution of genetic factors (12). To find out new prevention methods, possible mechanisms, and potential advanced therapies, we need to have a deeper understanding of the mutual interaction between VDR polymorphism and HCC. So far, there are several articles that have drawn our attention (25-27). However, the results have not been integrated yet; thus, we conducted a meta-analysis to give a more objective and compelling answer.
Our meta-analysis showed that Fok I polymorphism might increase the HBV-related HCC risks in the overall population in the codominant genetic model. It is inconsistent with the VDR gene polymorphism study conducted by Falleti et al. (28). The study concluded that the b/b genotype of BsmI and the T/T genotype of TaqI had a large possibility to be the risk factors, and they were significantly associated with the higher incidence of HCC. However, the cases and controls were liver cirrhosis patients caused by hepatitis B, hepatitis C, or alcohol. Thus, there were too many confounding factors to confirm the relationship between HBV and HCC. On the contrary, Huang et al. (29) pointed out that the development of HCC was not associated with VDR polymorphisms even though it could lead to different distinct clinical phenotypes in Taiwanese HBV carriers. Nevertheless, the Fok I polymorphism was not involved in the Taiwan study, which might have made the conclusion a little arbitrary.
The main objective of this meta-analysis was to amalgamate the individual studies and make a more powerful conclusion. However, some limitations need to be taken seriously into account. First, the numbers of included studies and patients are small. Besides, one of the three studies did not detect the other three VDR polymorphisms TaqI (rs731236), ApaI (rs7975232), and BsmI (rs1544410); thus, we had to focus only on Fok I (rs2225870). Therefore, our result might seem to be a little plain and subject to potential bias. Second, adjustments for some factors, which could influence the progression of HCC such as vitamin D levels, lifestyle (drinking or smoking), and environmental factors were not conducted due to limited available data.
The association of Fok I polymorphism with other diseases has been reported frequently. Sarkissyan et al. found that the Fok I polymorphism was associated with increased colorectal carcinoma (CRC) risks (30). Vogel et al. showed a strong relationship between F allele of the Fok I polymorphism and autoimmune hepatitis (31). Shafia et al. reported that the genetic variation of Fok I influences the susceptibility to and progression of myeloma (32). Our results in this analysis were consistent with the mentioned findings. However, the mechanisms of the relationship between the Fok I polymorphism and the mentioned diseases remain unclear. During the past two decades, additional functions of vitamin D have increasingly been demonstrated including antiproliferative, pro-differentiation, pro-apoptotic, anti-angiogenesis, and anti-invasive characteristics through many cancer cell experiments. Recent studies showed that when vitamin D binds to VDR, the expression of downstream P21 and P27 may decrease, which can inhibit proliferation and induce differentiation for various malignant cells through a G0/G1 phase arrest (33-35). The “f” allele tends to have less function than “F” allele and thus, may entail more risks for HCC. Interestingly, in a recent meta-analysis, He et al. reported that Fok I FF tends to be a risk factor for HBV infection, which is inconsistent with our results (36). The explanation was that the higher transcription activity of the F allele (C) seemed to inhibit the immune function of Th1 or Th2 and promote HBV infection (37), due to the molecular change of IL-12, IFN-γ, IL-4, and IL-10 (38, 39), finally affecting a series of subsequent pathways. The result indicated that the VDR polymorphism of people who were susceptible to HBV was different from the VDR polymorphisms of HBV-infected people who had a large probability to progress HCC; since the higher transcription activities initiated by VDR tend to be infected with HBV, lower transcription activities are more likely to be associated with HCC on the contrary. The difference seems to partly answer why only little HBV-infected patients finally progress to HCC.
In conclusion, we are one of the rare groups to provide a meta-analysis to evaluate the role of the VDR polymorphism on HCC risks in HBV patients. The “f” allele of the Fok I polymorphism was found to significantly increase the risk of HCC in HBV-infected patients. In the future, more studies with large sample sizes and more accurate analyses are needed to further confirm the conclusion.