1. Background
Even half a century after the development of the hepatitis B virus (HBV) vaccine, HBV infection is still a serious public health problem. HBV is recognized as one of the seven known human oncogenic viruses and is responsible for one of the highest yearly mortality rates (1). A high genome diversity allows HBV to evade the host immune system, develop resistant mutations against antiviral therapy (at P gene) and vaccination, and generate diagnosis-escaping variants (at the S gene) (2). Of note, naturally occurring mutations associated with drug resistance have been detected in treatment-naïve individuals and can be transmitted from person to person (3). These biological characteristics of HBV pose major challenges for the elimination of the virus. Therefore, HBV control is currently a priority to public health authorities. To this end, screening, vaccination, and monitoring of infected cases in high-risk groups is strongly recommended (4-6).
HBV is a blood borne virus and is transmitted through perinatal, sexual and parenteral/percutaneous routes. The high stability of HBV at room temperature (compared to other blood-borne viruses) (7), increases the chance of transmission through close contact in overcrowded places like prisons, refugee camps, and healthcare facilities (8-11). Furthermore, the risk of transmitting HBV is further enhanced by poor sanitation and aggressive behaviour in institutions for people with intellectual disability (12). Besides patients, also treating nurses/physicians are at risk of getting infected infection; especially people taking care of aggressive patients (13).
In Iran, the implementation of the national vaccination against HBV has significantly decreased the HBV prevalence (14). Recent epidemiological studies among the Iranian general population and blood donors reported the circulation of HBV genotype D in chronic carriers (14). Despite a few studies performed in high-risk groups, there is a lack of data characterizing the HBV strains circulating in persons with intellectual disability in Iran.
2. Objectives
In this study we applied molecular analysis to determine the seroprevalence of HBV in individuals with intellectual disability. For the first time, this report will provide valuable information of HBV infection among this vulnerable population in Iran.
3. Methods
This study was approved by the Ethics Committee of Shahid Beheshti University of Medical Sciences with approval number 25-1580-1393-122 and is in accordance with the Helsinki declaration of 1964. Individuals with intellectual disability from six different institutions in Tehran, Iran, were included in this study.
Inclusion criteria were defined based on census-based sampling. Informed consent was obtained from patient's parents or their guardian. Blood samples were taken after obtaining permission of the center director and based on the patient readiness for sampling. For instance, in some cases the patient mental state and aggressiveness could impede the sample collection. Individuals recruited in this study were semi-private and protected patients that suffered from mental and physical retardation. Institutionalized patients received treatment such as physiotherapy, occupational therapy and speech therapy for the intellectual disability patients. The patients had been admitted to the center by their families or without family supervision.
Blood samples were collected during 2011 - 13. Demographic data including age, sex, type of disability, period of visit to the centre, and HBV vaccination history were obtained through a questionnaire. HBV vaccination was reported by evaluating the patient files. If there was no record of HBV vaccination or incomplete vaccination, the patient was reported as HBV vaccination naive. If no vaccine card was available, the patient was recorded as unavailable data for vaccination.
HBV serological markers, including HBsAg and HBcAb, were determined for all patients, while HBsAb was only checked for HBV DNA positive cases (bioELISA-, Spain, Barcelona). Viral DNA was extracted from sera using the QIAmp® Viral DNA mini kit (QIAGEN, Hilden, Germany) in patients who were either HBsAg or HBcAb positive. To amplify the complete HBsAg gene, an in-house nested PCR technique was employed (15). Bi-directional sequencing was conducted using the ABI PRISM Big Dye® Terminator cycle sequencing reaction kit (Applied Biosystems, Foster City, CA, USA) on an ABI PRISM 3130 automated sequencer. Primer-walking was performed and a consensus sequence was generated by assembling the S gene segments using the SeqMan II® software (DNAStar Inc., Madison, Wisconsin, USA). Sequences were submitted to NCBI GenBank under accession numbers KY225828-KY225842. Possible recombination events were investigated using the SimPlot software, version 3.5.1. A nucleotide BLAST and a web-based HBV genotyping program were used to determine HBV genotypes. To confirm the genotyping results, a phylogenetic analysis was performed. HBV S gene sequences were extracted from NCBI GenBank and aligned using ClustalW, version 1.83. A neighbour joining tree was constructed under the Kimura 2-parameter substitution model using MEGA software V6. The reliability of the pairwise comparison and the phylogenetic tree analysis were assessed by bootstrapping using 1000 replicates. Genotypes were determined by comparing the full-length HBsAg to the NCBI reference sequences representing all HBV genotypes and subgenotypes. HBV subtypes were assigned based on the amino acid sequences of the surface antigen region. Data analysis was performed using SPSS (Ver. 17.0, Chicago, IL) and R software (version 2.10.1). Categorical data were compared using the chi-square or Fisher exact test as appropriate. Numerical variables were analysed using the independent sample t-tests. Statistical significance was considered at two-sided P < 0.05.
4. Results
Samples were collected from 215 (53.8%) males and 185 (46.3%) females who were hospitalized for the duration of 1 to 30 years (mean ± SD: 11.1 ± 4.9 years). The studied population included 293 (73%) persons with intellectual disability, 53 (13.0%) patients with Down’s syndrome, 42 (10.5%) patients with cerebral palsy (CP), and 13 (3.2%) patients with autism. The mean ± SD age of hospitalization was 4.53 ± 3.22 (0 - 13 years). HBV vaccination status was positive, negative, and unavailable in 194 (48.5%), 56 (14.0%), and 150 (37.5%) patients respectively (Table 1). Out of 400 samples, 28 (7.0%) and 41 (10.3%) were positive for HBsAg and HBcAb respectively. There was no statistically significant relationship between HBsAg/HBcAb positivity and age, gender, period of attendance, clinical manifestations, or vaccination history (P > 0.1) (Table 2). From 41 HBcAb-positive patients, HBV DNA was detected in 26 individuals, of which 20 patients were HBsAg positive and six HBsAg negative. There was no statistically significant relationship between HBV DNA detection and age, gender, period of attendance, or clinical history (all P > 0.1) (Table 2). HBV infection among different centers varied significantly (P < 0.01) between 2% - 17%.
| Centers | Patients in Each Center, N | Age, y (Mean ± SD) | Male, No. (%) | Female, No. (%) | HBsAg Positive, No. (%) | Anti-HBc Positive, No. (%) | Vaccination, No. (%) | Occult Infection, No. (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | 21 | 9.7 ± 2.5 | 14 (67) | 7 (33) | 2 (9) | 4 (19) | 21(100) | 2 (9.5) |
| 2 | 51 | 11.9 ± 2.9 | 23 (45) | 28 (55) | 6 (11) | 7 (14) | 37 (73) | 0 (0) |
| 3 | 99 | 11.3 ± 2.8 | 65 (66) | 32 (34) | 2 (2) | 7 (7) | 74 (75) | 1 (1) |
| 4 | 64 | 4.0 ± 2.3 | 25 (39) | 39 (61) | 8 (13) | 8 (13) | 48 (75) | 1 (2) |
| 5 | 144 | 12.6 ± 3.4 | 78 (54) | 66 (46) | 6 ( 4) | 10 (7) | 13 (9) | 1 (1) |
| 6 | 23 | 20.4 ± 5.6 | 10 (43) | 13 (57) | 4 (17) | 5 (22) | 1 (4) | 1 (4) |
| Laboratory Findings | Age, y | P | Gender (M/F) | P | Years of Attendance | P | Clinical Manifestation [Down, CP, MR, Otism], No. (%) | P | History of Vaccination, No. (%) | P |
|---|---|---|---|---|---|---|---|---|---|---|
| HBsAg positive | 11.18 ± 6.39 | 0.96 | 12/16 | 0.28 | 5.32 ± 4.17 | 0.18 | 4 (8), 7 (17), 20 (7), 1 (8) | 0.17 | 20 (8) | 0.6 |
| HBsAg negative | 11.14 ± 4.83 | 199/173 | 4.48 ± 3.13 | 49 (92), 35 (83), 276 (93),12 (92) | 230 (92) | |||||
| HBcAb positive | 11.76 ± 5.61 | 0.40 | 22/19 | 0.93 | 5.13 ± 4.11 | 0.19 | 5 (9), 9 (21), 30 (10), 1 (8) | 0.19 | 27 (11) | 0.3 |
| HBcAb negative | 11.08 ± 4.82 | 190/169 | 4.45 ± 3.09 | 48 (91), 33 (79), 266 (90), 12 (92) | 223 (89) | |||||
| HBV DNA + | 12.56 ± 6.03 | 0.13 | 12/14 | 0.41 | 5.89 ± 4.68 | 0.13 | 3 (6), 6 (14),16 (6),1 (8) | 0.17 | 18 (9) | 0.1 |
| HBV DNA - | 11.07 ± 4.80 | 204/170 | 4.44 ± 3.11 | 50 (94), 36 (86), 276 (94), 12 (92) | 232 (91) |
Abbreviations: CP, cerebral palsy; F, female; M, male; MR, mental retarded
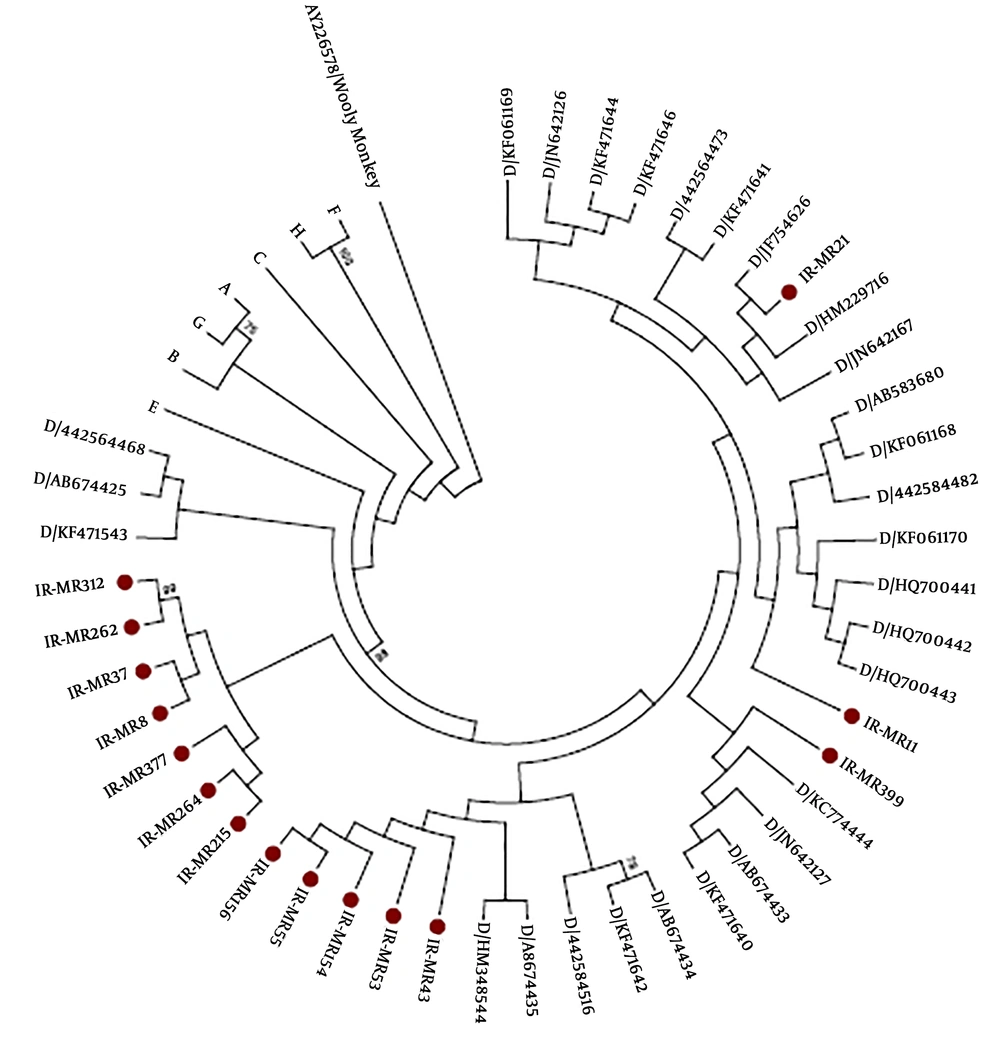
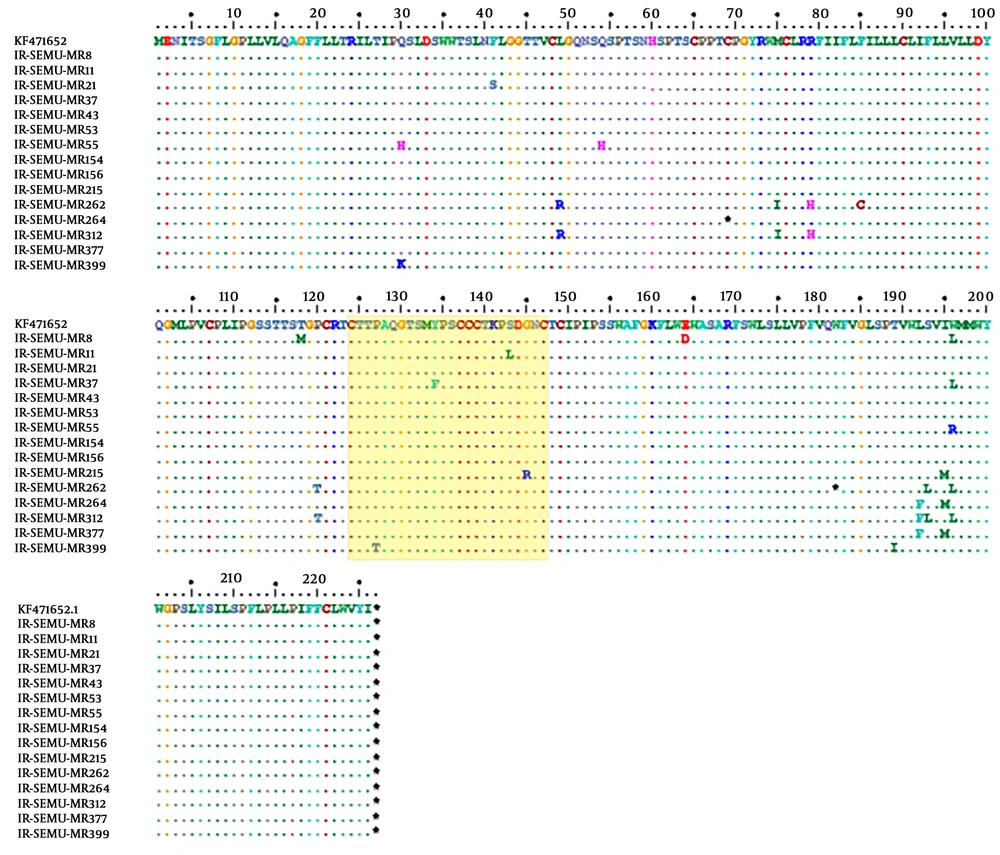
From 15 patients, partial S genes (full-length of HBsAg, 681 base pairs) were successfully sequenced and 11 out of 15 individuals showed anti-HBs antibodies in their sera (Table 3). Phylogenetic analysis of the 15 partial S genes and 72 strains derived from Genbank revealed that all identified strains belonged to genotype D (Figure 1). HBV subtypes were determined for all sequenced strains (n = 15) based on the amino acid motives in positions 122, 127, 140, 159, and 160 in the surface antigen region. This analysis presented two different types of HBV subtypes circulating in the studied population: ayw2 (n = 14, 93%) and ayw3 (n = 1, 7%) (Table 3). Although, no deletion, insertion, or recombination events were identified, several mutations were detected in the HBsAg region. Two stop codons were detected, one strain showed C69* while the other strain carried W182* at the HBsAg protein (Figure 2). The mutations, observed in the deduced HBsAg, have been depicted in Figure 2, and the medical importance of these mutations in the S gene has been explained in Table 4. The frequency and type of mutation in the HBsAg protein had no statistically significant relationships with the age or sex of the HBV-infected patients. Although the evaluation of resistance to antiviral therapy was not the main goal of this study, translation of the sequenced S gene into the ORF of the Pol gene revealed that eight out of 15 strains harboured antiviral resistance mutations in the reverse transcriptase domain of the polymerase gene. Surprisingly, none of these patients had any record of antiviral therapy.
Neighbor-joining phylogenetic tree based on complete HBsAg from 15 Iranian strains and 94 reference strains with different genotypes of HBV retrieved from the GenBank. A Woolly Monkey stain sequence (AY 226578) was used as an out-group. The complete HBsAg from this study is marked by red circle. HBV non- D branches were compressed. The numbers at the nodes represent the percentages of the bootstrap support values (1,000 replicates).
Distribution of amino acid mutations detected within the HBsAg among mentally retarded infected with HBV, the amino acid sequences represent positions at the 99-170 of the HBsAg is considered as Major Hydrophilic Region (MHR). The amino acid variations in the “a determinant” motif are framed in a yellow box.
| Patient ID | Gender | Age, y | Mental Disease Type | Center | Period of Attendance, y | HBV Vaccination History | HBsAg | Anti-HBsAg; Titer, mIU/mL* | Anti-HBcAg | HBV Genotype | HBV Subtype | Immune Escape Mutations | YMDD Mutation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MR8 | F | 12 | MR | 1 | 2 | Vaccinated | Pos | 5 | Pos | D1 | ayw2 | Pos | Pos |
| MR11 | M | 6 | MR | 1 | 1 | Vaccinated | Neg | 42 | Pos | D1 | ayw2 | Neg | Neg |
| MR21 | M | 11 | CP | 1 | 2 | Vaccinated | Neg | 2 | Pos | D1 | ayw2 | Pos | Neg |
| MR37 | F | 13 | CP | 2 | 3 | Vaccinated | Pos | 52 | Pos | D1 | ayw2 | Pos | Pos |
| MR43 | F | 14 | CP | 2 | 11 | Vaccinated | Pos | 461 | Pos | D1 | ayw2 | Neg | Neg |
| MR53 | F | 14 | Down | 2 | 11 | Vaccinated | Pos | 48 | Pos | D1 | ayw2 | Neg | Neg |
| MR55 | F | 14 | CP | 2 | 10 | Vaccinated | Pos | 407 | Pos | D1 | ayw2 | Pos | Neg |
| MR154 | M | 13 | MR | 3 | 2 | Vaccinated | Neg | 277 | Pos | D1 | ayw2 | Neg | Neg |
| MR156 | M | 9 | Au | 3 | 3 | Vaccinated | Pos | 24 | Pos | D1 | ayw2 | Neg | Neg |
| MR215 | F | 4 | MR | 4 | 1 | Vaccinated | Pos | 362 | Pos | D1 | ayw2 | Pos | Pos |
| MR262 | M | 21 | MR | 6 | 13 | No history | Neg | 4 | Pos | D1 | ayw2 | Pos | Pos |
| MR264 | F | 30 | MR | 6 | 13 | No history | Pos | 6 | Pos | D1 | ayw2 | Neg | Neg |
| MR312 | M | 5 | MR | 5 | 0 | No history | Pos | 15 | Pos | D1 | ayw2 | Pos | Pos |
| MR377 | M | 11 | MR | 5 | 5 | No history | Pos | 276 | Pos | D1 | ayw2 | Pos | Pos |
| MR399 | F | 16 | MR | 5 | 10 | No history | Pos | 11 | Pos | D1 | ayw3 | Pos | Neg |
| Position at the HBsAg | Wild-Type Amino Acid | Mutant Amino Acid | Position at “a Determinant” | Affect Serological Diagnosis | Vaccine Escape | Mutant Failure in HBIg Therapy | References |
|---|---|---|---|---|---|---|---|
| 30 | Glutamine | Histidine | No | ND | ND | ND | |
| 41 | Phenylalanine | Serine | No | Yes | ND | ND | (16) |
| 54 | Glutamine | Histidine | No | ND | ND | ND | |
| 69 | Cysteine | Stop codon | No | ND | ND | ND | (17, 18) |
| 118 | Threonine | Methionine | Yes | Yes | ND | ND | (19) |
| 145 | Glutamine | Arginine | Yes | Yes | Yes | Yes | (20) |
| 164 | Glutamate | Aspartate | Yes | ND | Yes | Yes | (21) |
| 182 | Tryptophan | Stop codon | No | ND | No | No | |
| 195 | Isoleucine | Methionine | No | ND | Yes | Yes | (21, 22) |
| 196 | Tryptophan | Leucine | No | No | Yes | No | (21, 23) |
| 196 | Tryptophan | Arginine | No | ND | ND | ND |
Abbreviation: ND, non-determined
5. Discussion
In this study, a higher prevalence of HBsAg (7%) was found in institutionalized patients with mental disability compared to the general population. Also, this number is higher than the HBV prevalence among high risk group (drug users, sex workers and prisoners) that is estimated at 4.8% (95% confidence interval: 3.6% - 6.1%) in Iran.
In contrast, HBcAb was detected in 10.3% of the study population, which is lower than the general population (24). The high seroprevalence of HBsAg found in our study (7%) corresponds to previous reports of HBV infection in disabled children (8%) from Iran in 2003 (25). Considering the mean age of the study population (11 years), most patients were born after the implementation of the national vaccination programme for neonates in Iran in 1993. Of note, we were unable to obtain vaccination data for 37% of the study population due to missing vaccination cards, which could have an impact on the absolute vaccination coverage. For instance, some patients were homeless at the moment of hospitalization and no vaccination data could be retrieved. The high prevalence of HBsAg could be related to the reduced immunocompetence of these patients (26). However, a recent study rejected this hypothesis and demonstrated a high efficiency of the HBV vaccine in this group of patients (27). Based on our findings, we hypothesize that some patients were infected via mother-to-child transmission (MTCT) and that infecting strains were shared with other patients via horizontal transmission. Despite HBV vaccination and immunoglobulin administration at the time of birth, immunotherapy prophylaxis can fail if the mother is HBeAg positive and has a high viral load. The detection of several (73%) vaccine/immuno escape mutants coexisting with HBs antibody supports this hypothesis in our study. In addition, the identical mutational pattern of the isolates and the identification of several occult cases (six out of 15) corroborate this idea. Likewise, the topology of the phylogenetic tree presents a signature of sharing a common ancestor of circulating strains among these patients. Particular behavioural characteristics of retarded patients such as increased incidences of bleeding, fissured or hypertrophied gums or lips, self-mutilation with bleeding wounds, excessive drooling or spitting, and scratches or biting may facilitate intra-group viral transmission (12). In literature, transmission of HBV (wild or mutans strains) through close contacts has been documented. For instance, a study performed in the USA (28) reported the transmission of HBV and other blood borne viruses (e.g. HIV) among mentally ill residents at a long term care facility. Confirming horizontal transmission events requires meticulous phylogenetic analysis by using HBV full-length genomes (7). However, the low sample volume/availability and low viral loads restricted this analysis. HBV infection among different centers varied significantly (P < 0.01) between 2% - 17%. We could not find any possible cause for this marked difference based on our available data.
The detection of HBV genotype D is in line with previous epidemiological studies in Iran (29, 30) and the neighbouring countries (8, 31, 32). Although HBV D1 is the major subgenotype circulating in Iran (33), partial S gene sequences in the present study did not provide enough data for HBV subgenotyping (34). Previously, it has been shown that HBV subtypes ayw2 and ayw3, which were identified in this study, are associated with subgenotype D1 (35). In addition to the epidemiological role of HBV subtypes, it has been shown that ‘ay’ has less reactivity than ‘ad’ in ELISA assays and is accompanied by a lower vaccine response (36).
Single or combined mutations in HBsAg were detected in 73% of the isolates. These mutations can modulate the structure of the HBsAg molecule and help the virus to escape vaccine, immunotherapy, or diagnostic assays (37). In this study, the frequency of naturally occurring MHR mutations was 53.3%, indicating that a significant number of strains carry one or more mutations in this antigenic motif (P < 0.05). This rate is much higher than the numbers reported by previous studies involving blood donors (32.8%) (3) and the general Iranian population (17.2%) (38). Noteworthy, half of the mutations in MHR were located in the ‘a determinant’ region, which is also higher compared to previous studies from Iran (3).
In this study, codon 120 was most frequently mutated. This finding complies with previous studies from Iran (39), Morocco (40), and Serbia (41), where HBV genotype D is the predominant circulating strain. This mutation is mostly detected with HBs antibodies (42), and is responsible for most diagnostic and vaccine escape mutants in the HBV S gene (43). Moreover, we identified a variation in codon 145 (G145R) in one strain. Both mutations (P120T and G145R) are well known as vaccine, immunoglobulin escape causing mutations that can emerge under immunotherapy or antiviral therapies (44). Both can be transmitted vertically and horizontally.
Both upstream and downstream premature stop codons of the S gene can lead to truncated proteins and failure in the HBsAg diagnostic assays (45). Two strains with premature stop codons upstream (sC69*) and downstream (sW182*) were identified in the present study. These mutations were previously reported in Iranian blood donors and cirrhotic patients (17). Surprisingly, one of those strains carried P120T along with sW182* and was presented as HBsAg negative in the serological assay. However, there was no significant correlation between identified mutations in isolated strains from occult or overt cases.
The development of antiviral resistance mutations in HBV polymerase gene can have major clinical consequences. Furthermore, overlap of the polymerase and surface gene leads to variations that affect invasiveness, vaccine efficacy, and the intra-population transmission of the virus. The detection of antiviral resistance in treatment naïve patients that harbour viruses with these mutations increases the public health concern (3). In the present study, eight out of 15 strains developed single mutations associated with Lamivudine resistance (rtM204V/I). In seven strains these mutations were accompanied by rtL180M mutation, and in one strain together with rtL180M, rtV173L, and rtL80I. Surprisingly, none of our patients had a history of antiviral treatment. This rate (more than 50%) of antiviral resistance mutations in treatment naïve patients is much higher than previously reported (46, 47). This finding highlights the possibility of intra-group transmission of drug resistant variants. We hypothesize that some cases were born to HBsAg-positive mother(s) who were under antiviral therapy and had developed antiviral resistance before delivery. These resistant strains were vertically transmitted to the new-borns. Subsequently, close contact of children in healthcare institutions provided the setting for horizontal transmission. Although antiviral resistance mutations in treatment naïve patients have already been reported in Iran, the present study is the first report of horizontal transmission and with such a high rate. In line with our findings, several reports have demonstrated the transmission of HBV antiviral resistance mutations in high-risk cohorts (48, 49).
Of note, despite the importance of early and complete HBV vaccination of all health care workers, nurses are still at significant risk of developing needle stick injuries and mucocutaneous exposure. Therefore, a continuous training program to raise awareness of viral blood borne infection risks is a necessity among nurses. The knowledge about the nature of HBV disease, prevention, treatment and vaccine availability was unsatisfactory in different studies (50). This limited knowledge among health care workers may increase unsafe practices in the institutions (51).
One important finding in this study was that 81% of HBsAg positive patients were also positive for HBsAb. HBsAg coexistence with HBsAb has been reported previously among chronic hepatitis B patients with HBsAg mutants. However, in the present study 60% of patients, who were positive for HBsAg and HBsAb, were infected by immune escaped mutations. Researchers in 2006 and 2007 (52, 53) reported that in CHB patients, the coexistence of HBsAg and HBsAb was associated with an increase of “a” determinant variability (region between amino acid residues 99 and 169 which are involved in the binding of antibodies against HBsAg). Based on this observation they suggested a selection of HBV immune escape mutants during chronic carriage stage (52). The consequences of this selection process with regard to vaccine efficacy, diagnosis, and clinical evolution remain partially unknown. In our study, all centers directors mentioned that all institutionalized patients were regularly screened based on HBsAb titer. It can be concluded that if only HBsAb test is used for HBV screening, HBV positive cases can be missed in mentally retarded centers. Furthermore, we observed a high prevalence of mutations in “a determinant” region and immune escaped mutations in HBsAg and HBsAb positive patients.
In conclusion, residing and working in crowded places with the possibility of close contact has led to the spread of HBV and can be a potential reservoir for the dissemination of HBV. This epidemiology of HBV in institutions for persons with intellectual disability, calls for the need of comprehensive and national studies in similar establishments. These studies could have a key role in controlling HBV in the post-vaccination period.