1. Background
After lung and breast cancers, hepatocellular carcinoma (HCC) is currently the third cause of cancer mortality in humans. The prognosis of the disease remains particularly dismal with an overall mortality that corresponds to 95% of its incidence (1). According to the World Health Organization, in the great Middle East and North Africa (MENA) region severe liver diseases, including HCC, are among the pathologies that have grown in prevalence during the last decades (2).
In Tunisia, several HCC risk factors are significantly prevalent in the general population. Indeed, persistent infections with hepatitis B (HBV) and C viruses (HCV) represent important public health issues for the country with historically high endemic levels of HBV carriage in Southern Tunisia, while HCV has presumably spread more recently essentially through iatrogenic transmission around selected urban centers of northern regions (3, 4). More recently, the large increase in the non-alcoholic fatty liver disease (NAFLD) cases has been identified as a significant contributor to an insidious degradation of liver health in Tunisia (5).
Besides infectious and metabolic risk factors, environmental agents such as harmful mycotoxins have been suspected for decades to play a significant role in the epidemiology of liver diseases observed in Tunisia (6). Most noticeably, Tunisian researchers have consistently isolated aflatoxin B1 (AFB1), a major liver carcinogen, in local food components of vegetal (maize, sorghum, nuts) or animal (milk) origins (7, 8).
Furthermore, a few years ago, Public Health authorities issued a warning regarding the consumption of AFB1-contaminated nuts from Alleppo pine (zgougou in local language, Pinus halepensis), a major component of some of the most popular Tunisian pastries. Despite the considerable concerns generated by this alarming information, no molecular data measured the consequences of AFB1 exposure in patients with liver diseases. For this reason, we set out to explore the free circulating DNA (fcDNA) from Tunisian patients with HCC for the presence of the AFB1-induced TP53 mutation at codon 249 (ARG249SER) (9). This G>T mutation affects codon 747 of TP53 and has been consistently associated with AFB1 exposure in all geo-epidemiological backgrounds tested so far (10, 11). In addition, we looked for the presence of the TERT promoter mutation at nucleotide-124 that is considered as the most prevalent somatic alteration in human HCC (12).
To increase the probability of detecting minute amounts of mutants, we decided to use the droplet digital PCR (ddPCR) technique. ddPCR technique allows the independent amplification of microcompartments and their subsequent one-by-one analysis in a microfluidic device through the partitioning of reagents in 15,000 - 20,000 pico-droplets. Although its use is not recommended in all circumstances, it has been recurrently shown in the recent years either for cancer mutations monitoring, infectious agent detection, or prenatal diagnosis that ddPCR is more sensitive than traditional quantitative real-time PCR to detect targets that represent only a small fraction of a much larger and more complex population of DNA molecules (13-17).
2. Methods
2.1. Patients
This case-control study was performed among HCC patients referred to Gastroenterology departments of La Rabta (Tunis) and Tahar Maamouri hospitals (Nabeul) and the Department of Surgery of Mongi Slim Hospital (La Marsa, Tunis). The diagnosis of 47 HCC cases was based on clinical symptoms and the status of the non-tumor liver, presence of a liver mass at ultrasound, the serological context, hematoxyllin-eosin staining of tissue section and measurement of serum alpha-fetoprotein (AFP) levels. Cases were individually 1:1 paired-matched by sex and age (± 5 years) with control subjects consecutively selected and represented by patients with or without serological signs of chronic liver infection presenting to the same medical departments during the same period. Investigations were approved by the Ethics Committee of Tunisian Ministry of Health. An informed consent was obtained for each patient enrolled in the study.
2.2. DNA Extraction and Quantification
Sera were collected, aliquoted and stored at -80°C. Circulating free DNA was extracted from a mean volume of 600 µL of serum. Briefly, sera were digested overnight at 37°C under mild agitation in a buffer corresponding to a final concentration of 10 mM EDTA, 2% sodium dodecyl sulfate (SDS) and 0.1 µg/mL proteinase K (Eurobio, Les Ulis, France). Samples were then successively submitted to a phenol pH 8.0 and to chloroform-isoamylalcohol extractions. Aqueous phase was precipitated by ethanol (2.5 volumes) overnight at -80°C in the presence of 0.1 of 3 M sodium acetate, pH 6.0 and T40 dextran (2 µL). After 60 min/14000 rpm centrifugation, DNA pellet was rinsed in 70% ethanol at room temperature, solubilized in TE pH 8.0 buffer and quantified by Qubit dsDNA HS assay kit (Life Technologies, Villebon, France).
2.3. Circulating Mutations Detection
Amplifiability of cfDNA was first checked on a single copy nuclear gene (MGEA5, meningioma expressed antigen 5, sense primer, CCCGTACAAAGGAAGATCCA, reverse primer, ATTTGTACCAGGTGGCAAGG) by a standard quantitative real-time PCR. Samples that yielded satisfying results were further studied. The assays employed were a TaqMan™ SNP Genotyping Assay (C-60493487-10, Life technologies, Villebon, France) for TP53 R249S and the PrimePCR™ ddPCR™ Copy Number Assay (dHsaCP1000100, Bio-Rad, Marnes-la-Coquette, France). Droplet digital PCR reactions were performed on the QX100 system (Bio-Rad, Hercules, CA, USA). Reaction mixture consisted of10 μl ddPCR Supermix (Bio-Rad, Marnes-la-Coquette, France), 1.6X TaqMan™ SNP Genotyping Assay (C-60493487-10, Life technologies, Villebon, France), and 3 µL of cfDNA isolated from plasma samples in a final volume of 20 µL was mixed with 70 µL of droplet generation oil (Bio-Rad) and partitioned into approximately 20,000 droplets in the QX100 droplet generator (Bio-Rad). The droplets generated from each sample were transferred to a 96-well plate and PCR amplification was conducted in an iCycler PCR instrument (Bio-Rad) with the following conditions: 95°C for 10 minutes, 40 cycles of 94°C for 30 sec with a ramping of 2°/sec, 59°C for 1 minutes with a ramping of 2°/sec, followed by 98°C for 5 minutes and a hold at 4°C. After amplification, the 96-well plate was loaded onto the QX100 droplet reader (Bio-Rad) that automatically measures the fluorescence intensity in individual droplets. The generated data were subsequently analyzed with QuantaSoft™ software (Bio-Rad) based on positive and negative droplet populations. For TP53, genomic DNA from the PLC/PRF5 HCC cell line that harbors the R249S mutation was included as a positive control in each experiment, while a genomic DNA from a tumor mutated in -124 of TERT promoter was used as positive control. The genomic DNA extracted from peripheral blood mononuclear cells of a subject without any mutants was used as negative control in all the experiments.
Samples positive for TP53 R249S or TERT -124C>T were tested at least twice in two independent runs, while samples negative for mutants but yielding more than 1000 droplets for the control locus MGEA5 where tested once and scored as negative. Samples negative for mutants and displaying less than 1000 droplets positive for control locus were tested at least twice in two independent runs. Samples discarded from the analysis did not generate any fluorescent droplets (one sample for patients and controls) or a sufficient merged number of droplets positive for the reference gene MGEA5. Samples were considered positive for TP53 R249S or TERT -124 c>t when at least two droplets were found positive for mutant alleles in two independent droplet digital PCR runs.
2.4. Statistical Analyses
Statistical analyses were performed using Prism 6.0 statistical package. Numerical variables were summarized by their median, mean and range according to their types of distribution (normal or not). They were compared either by a Student’s t-test or by Mann-Whitney test as appropriate. Categorical variables were summarized as frequencies that were compared by Fisher’s exact test. All the tests were two-sided and the level of significance was set at P < 0.05.
3. Results
The clinico-biological characteristics of the patients are described in Table 1. Patients and controls were essentially different in terms of the seroprevalence of anti-HCV and non-infectious risk factors for HCC. Circulating free DNA (cfDNA) concentrations in sera were similar in patients and controls (~ 480 ng/mL).
| Clinico-Biological Features | HCC, N = 47 | Controls, N = 51 | P Value |
|---|---|---|---|
| Age | 62.2 ± 9.3 | 63.7 ± 11.5 | ns |
| Sex ratio | 0.95 (23:24) | 0.82 (23:28) | ns |
| Serological features, % | |||
| HBsAg | 23.4 | 9.8 | 0.10 (ns) |
| Anti-HCV | 53.2 | 5.9 | 2.3E-07 |
| Anti-HBc | 55.3 | 37.2 | 0.10 (ns) |
| Anti-HBs | 19.1 | 35.3 | 0.11 (ns) |
| HBeAg | 8.5 | - | - |
| Anti-HBe | 38.3 | - | - |
| Anti-Delta | 2.1 | 0.0 | ns |
| NonB-nonC | 25.5 | 84.3 | 4.0E-09 |
| Other risk factors, % | |||
| Tobacco consumption | 34.7 | 13.9 | 0.02 |
| Alcohol intake | 24.4 | 0.0 | 4.8E-04 |
| Type 2 diabetes | 31.1 | 6.6 | 0.002 |
| AFP, ng/mL, % | |||
| > 300 | 33.3 | - | - |
| 10 - 300 | 38.4 | - | - |
| < 10 | 28.2 | - | - |
| Liver cirrhosis | 76.5 | - | - |
| Child-pugh stage, % | |||
| A | 35 | - | - |
| B | 56 | - | - |
| C | 9 | - | - |
| Circulating free DNA, ng/mL | 484.2 ± 81.8 | 489.1 ± 45.7 | ns |
| Prothrombin time, % | 68.2± 13.6 | - | - |
| Platelets, /mL | 115 ± 66 | - | - |
| Tumor diameter, main nodule, mm | 41 ± 29 | - | - |
| Multiple nodules, % | 52.2 | - | - |
| Aspartate aminotransferase, IU/mL | 82 ± 61 | 51 ± 59 | 0.018 |
| Alanine aminotransferase, IU/mL | 70 ± 58 | 59 ± 80 | 0.47 (ns) |
| γ-Glutamyl transpeptidase, IU/mL | 130 ± 140 | 28 ± 32 | 0.0069 |
| Alkaline phosphatases, IU/mL | 377 ± 787 | 153 ± 134 | 0.10 (ns) |
| Total bilirubin, mg/L | 31 ± 35 | 26 ± 50 | ns |
Demographic, Clinical and Biological Features of Patients with Hepatocellular Carcinoma and Controls
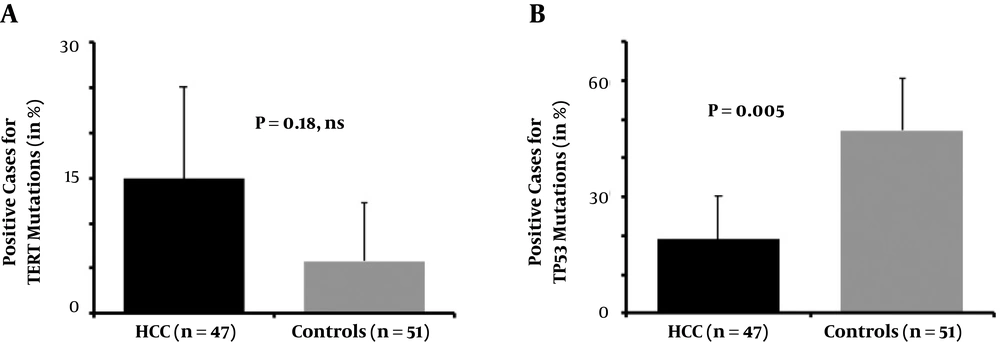
From the initial series, 47 and 51 serum samples collected from patients with HCC and from controls yielded a number of droplets suitable for further interpretation of the mutants searched. For TERT mutants, a median of 1950 droplets (range: 610 - 23761) was analyzed. There was no significant difference in droplet numbers between HCC and controls. We found that seven patients with HCC harbored a -124 C>T TERT mutation (n = 7/47, 14.8%), whereas three control participants were carrying this mutation (n = 3/51, 5.8%, OR = 2.7, 95% CI: 0.58 - 7.6, P = 0.18, ns, Figure 1A). The ratio of TERT-mutated positive droplets for patients with HCC ranged from 1.0E-03 to 8.3E-03 and the corresponding value of the control subject was similar (4.3E-03).
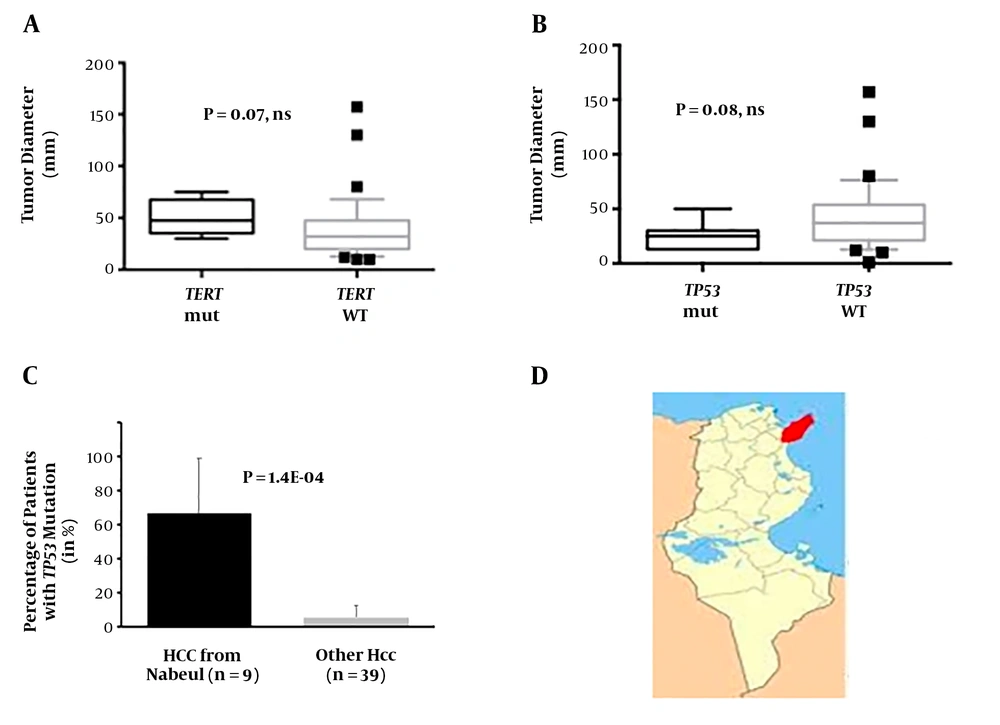
Patients with TERT mutations were marginally different from the rest of the series. All of them were presenting with antecedents of persistent infection with a hepatitis virus. Indeed, six of them were seropositive for anti-HCV, and the last one was carrier of HBV surface antigen (HBsAg). None of them was nonBnonC, while a sizeable proportion of the patients with wild-type TERT were (0.0% vs. 28.4%; P = 0.166, ns). Liver injury tends to be more important in TERT-mutated patients as shown by alanine amino-transferase levels (110 vs. 63 IU/mL; P = 0.072, ns). All the patients with TERT mutant had a cirrhotic liver, a situation in keeping with the pathophysiology of this molecular alteration that appears mostly in regenerative nodules in strongly fibrotic liver tissues. In three patients presenting this mutant, AFP levels were below the common diagnostic threshold for HCC (< 300 ng/mL) suggesting that search for TERT mutants might be useful to complement diagnostic armamentum for some patients with HCC. The size of the tumors in TERT-positive patients tended to be larger than those with the wild-type version (Figure 2A). Patients with TERT mutants were coming from only three regions from Northern Tunisia (Jendouba, Nabeul and Tunis governorates).
A, Tumor diameter in mm of the patients with detectable circulating TERT mutants and in those without detectable mutants. B, Tumor diameter in mm of the patients with detectable circulating TP53 mutants and in those without detectable mutants. C, Proportion of patients originating from Nabeul and the rest of country with detectable circulating TP53 mutants. D, Location of Nabeul governorate in Tunisia.
Concerning the circulating TP53 mutants, the situation was drastically different. A subset of nine patients with HCC (n = 9/47, 19.1%) was positive for TP53 R249S mutant, whereas in controls this rate was surprisingly higher (n = 24/51, 47.0%; P = 0.005; Figure 1B). In HCC patients, 6.0E-04 to 2.7% of the droplets were positive (analyzed droplet median = 2009, range: 606 - 11050), whereas in controls between 6.0E-04 and 4.1% were loaded with a mutant version of codon 249 (median = 2401, range: 627 - 20728; P = 0.23, ns). Two patients with R249S were also carrying TERT-124 C>T, whereas it was also the case for a single control subject.
Clinical features of the patients with the circulating R249S were not drastically different from those observed in the remaining cohort. Among the differences, though, none of them was affected with type 2 diabetes, whereas it was the case in 36.8% of the R249S-negative patients (P = 0.041). Interestingly, tumor sizes of mutant carriers were, albeit non-significantly, rather small (25.8 ± 4.8 mm vs. 43.0 ± 5.1 mm; P = 0.082, ns; Figure 2B) suggesting that ddPCR detection of this mutation might take place early in tumor surveillance. The most remarkable feature of the TP53 R249S-positive HCC patients was their origin as six out of nine (66.6%) were coming from Nabeul governorate, whereas 2 out of 38 R249S-negative patients were coming from this governorate (5.2%; P = 1.4E-04, OR = 32.3, 95% CI = 3.9 - 468.0; Figure 2C). This observation suggests that the impact of AFB1 on liver health in Tunisia might be heavier in the northern coastal region of Nabeul (Figure 2D).
Control subjects positive for R249S mutations were not different from other controls in any of the available demographic, clinical or biological features. Overall, our analysis indicates that an important proportion of the Tunisian population, either affected by a serious liver disease or with an apparently healthy liver, is exposed to the mutagenic activity of AFB1. Controls with R249S were coming from eight different governorates with Tunis (n = 9) and Jendouba, (n = 6) as main geographical origins.
4. Discussion
Due to an historically large diffusion of some of its main risk factors (e.g., HBV and alcohol intake) and the recent spread of novel etiologies (e.g., HCV and fatty liver diseases), HCC incidence has progressed in most regions of the world in the last decades and so did the corresponding mortality. In the immense majority of cases, HCC is secondary to a protracted liver disease that frequently progressed to cirrhosis explaining why in most cases patients present to tertiary healthcare institutions with an already significantly altered personal clinical status. The prognosis of the disease is thus overwhelmingly poor, especially in low- and middle-income countries where the surveillance system is usually not adequate. Implementation of new biomarkers capable that can be combined with alpha-fœtoprotein (AFP) detection to diagnose early forms of HCC is thus of paramount importance to improve the rate of cure or to reasonably extend the median survival rate of patients with HCC. High-throughput sequencing or new forms of PCR techniques currently provide the possibility to explore the blood for the presence of rare molecules shed by tumor cells in an approach conveniently designated as liquid biopsy.
In the great MENA region defined by the World Health Organization, the overall prevalence of chronic infections with hepatitis B (HBV) or C (HCV) viruses ranges around 2% - 3% but reaches 15% - 20% in Egypt (i.e., between 22 - 26 million individuals are concerned) (18-20). In addition, MENA is known as one of the regions (with the United States of America) with the highest rates of overweight/obesity, metabolic syndrome, and type-2 diabetes (T2D) (21). Indeed, metabolic disorders are currently affecting MENA populations as early as infancy or childhood, a situation predicted to have deleterious health consequences for the future grown-up generations. Dysmetabolic conditions have emerged recently as a major cause of morbidity for the liver and they represent important risk factors for terminal diseases such as cirrhosis and HCC (22, 23). The outcomes of such worrying epidemiological situation are that liver cirrhosis represents currently the 6th cause of death in the region and HCC incidence had massively increased in the past decades (2). In Tunisia, itself, with a little bit more than 110 cases of HCC per year, the toll taken by primary liver cancer is thus considered as relatively mild according to GLOBOCAN 2012 (1). However, it has been frequently suggested and recently substantiated that in low- and middle-income countries (LMICs) the incidence of HCC is probably significantly underestimated (24). In addition, in Tunisia, the median survival time of patients diagnosed with HCC is low as it barely exceeds 6 months (25).
Recent works have shown that -124C>T mutation in TERT promoter represents undoubtedly a very promising DNA marker for early HCC detection (12). In the present series, this mutant was found in almost 15% of patients but also in 6% of controls, thereby precluding any significant association with liver tumorigenesis due to the small size of the tested groups. However, in accordance with data from whole genome sequencing, we observed that the TERT point mutation was more abundant in HCV-infected patients (22.2%) than in HBV-infected cases (10.0%) or uninfected patients (0.0%) (26). In HBV-infected patients, a significant proportion of TERT alterations are actually provided by HBV DNA integrations in the gene promoter making point mutant no longer necessary for tumorigenesis. We consider, therefore, that -124C>T mutant still represents a promising biomarker in the monitoring of cirrhotic patients at risk for HCC, or the diagnosis of HCC in case of ambiguous imaging assessment of the liver nodule.
Global climate warming is suspected to extend the areas where aflatoxin-producing strains of Aspergillus will be distributed (27). As an example, sterigmatocystin, a precursor metabolite of aflatoxin B1, has been retrieved recently in grains from Norway (28). The long-time proven presence of AFB1 in a significant proportion of staple food consumed in Tunisia joint to the possibility to detect its mutagenic consequences in the blood of patients with HCC make Tunisians an attractive target to test the presence of tumor-associated alterations in the blood (7, 8, 29-34). The present work confirms the warning issued for decades by environmental scientists and proved that Tunisians are or have been recently exposed to the mutagenic activity of AFB1.
Our data suggest that Aleppo pine nuts are probably not the principal culprit in this situation. This nut is produced mostly in the central mountains in the region of Kasserine and consumed throughout the country, whereas TP53 R249S is found mostly in patients from Nabeul governorate on the north eastern coast of Tunisia. The Nabeul governorate is a major agricultural region in Tunisia with large segments of irrigated cultures dedicated to the production of oranges (Citrus reticulata) or chili pepper (Capsicum), the latter used in the national condiment Harissa. The spoilage of oranges (i.e., fruits and juice) or chili pepper with Aspergillus and their subsequent contamination by aflatoxin have been already described (35, 36). Our results should represent a strong incentive to conduct field studies in Nabeul governorate and elsewhere in the country with the aim of finding the eventual source of AFB1.
Among the questions raised by our analyses is the counter-intuitively higher prevalence of TP53 R249S mutants in controls than in patients with HCC. A logical explanation could be that the number target cells (i.e., hepatocytes) is much higher in healthy subjects than in patients with HCC who are generally affected with liver cirrhosis. In such circumstances, the parenchyma is replaced by fibrotic scars that lead on the long term to liver failure characterized by a number of hepatocytes insufficient to produce enough clotting factors or to detoxify ammonia.
In addition, liver cirrhosis is associated with a reduction of blood flow in the liver materialized by collateral venous circulation. Taken together, these phenomena might both contribute to reduction in the number of liver targets for AFB1 and the access of the mycotoxin to its primary site of action. In our opinion, this pathophysiological process might explain the higher prevalence of TP53 R249S in subjects with a healthy liver when compared to patients with a terminal liver disease. Two reports from Egypt have explored the presence of TP53 R249S mutants in cfDNA of patients with HCC. Hosny et al. (37) found a higher prevalence of mutation carriers in patients without HCC but with HBV infection (16.7% vs. 1.3%), whereas El-Din et al. (38)detected more mutants in patients with HCC than in controls with liver cirrhosis (15.0% vs. 0.0%). Overall, these results are consistent with our hypothesis that considers cirrhotic livers as cell-poor tissues unfavorable to the constant release of the R249S mutation and to its subsequent detection in plasma. In keeping with this hypothesis, Traore et al. analyzed a population of HBV-infected subjects in Bamako (Mali, South of the Sahara) and found more than 40 R249S copies/mL of plasma in 60% of cases (39). The use of ddPCR as a quantitative technique might, however, enable us to sort-out in the future between an ongoing clonal cell proliferation indicative of a tumor process and the presence in the blood of genomes from isolated cells killed directly either by aflatoxin B1 or by immune cells chasing for neoantigens generated by AFB1 mutagenesis.
As suggested previously, it is highly plausible that in absence of concomitant persistent infection with a hepatitis virus, control subjects with healthy liver will not develop HCC despite the presence of R249S mutant in their sera (37). In these populations, TP53 R249S is thus not a bona fide tumor biomarker but merely a marker of exposure to AFB1. Beyond the issue of HCC that is supposedly of minor concern for public health in Tunisia, our work implies that AFB1 might produce in Tunisia its deleterious effects outside the liver on various segments of the population. It is well-known that besides its widely advertised mutagenic activity, AFB1 is endowed with multiple toxicities that affect most notably the growth of children and the immune system of the subjects exposed (40-42). These potential extra-hepatic consequences of AFB1 activity on the Tunisian population might thus represent an ominous challenge for public health.
Our study suffers from an important limitation, that is, the small number of patients investigated. This shortcoming prevents any firm and definitive conclusion concerning the true distribution and in vivo impact of AFB1 contamination on the Tunisian population. Our work is, however, seminal as it is the first to be conducted on that field in the whole Maghrib region that consists of more than 86 million habitants. It should be therefore considered as a whistleblowing study aiming to stimulate more ambitious research programs and to attract the attention of public health stakeholders.
In conclusion, our study indicates that droplet digital PCR might represent a useful tool to complement the current procedures of HCC diagnosis. In addition, our data indicates that this technique could be useful when implemented in public health research. In the present case, it seems that middle-aged Tunisians are heavily exposed to the mutagenic activity of AFB1 as previously suggested by environmental and food scientists (6). Urgent initiatives are now necessary to confirm our data and eventually reduce the impact of AFB1 on Tunisians, as its deleterious activity might extend well beyond the carcinogenic risk, and inter alia, impacts with unforeseeable longer-term consequences the health of the younger strata of the population.