1. Background
Nonalcoholic fatty liver disease (NAFLD) is the most abundant chronic liver disorder (1), affecting 6.3% - 33% of the population worldwide (2). The prevalence of NAFLD is nearly 12–24% among Asian countries (3). Variety of NAFLD incidence may be attributed to the differences in age, gender, ethnicity, location, genetic effects, and lifestyle modification (4, 5). Studies showed that NAFLD may progress to both cirrhosis and hepatocellular carcinoma (HCC) (6, 7). Therefore, early diagnosis of NAFLD may be important to prevent these disorders. Diagnostic approaches for detection of NAFLD include serum liver enzyme measurement, imaging analysis such as sonography and magnetic resonance imagining (MRI) and liver biopsy (8). The last one is the gold standard for diagnosis of NAFLD, but this method is invasive and expensive and lots of subjects with suspected NAFLD are reluctant to use it as a diagnostic approach. Other traditional non-invasive methods mentioned above are not precise and sometimes cannot detect the patients with NAFLD (9). Thus, these challenges motivated researchers to find a non-invasive method with high accuracy for early detection of NAFLD.
In the recent decades, “omics” technologies such as metabolomics, the newest member in the “omics” approaches have provided an opportunity to find diagnostic biomarkers, clarify the underling mechanisms, and evaluate the treatment of various diseases using the nuclear magnetic resonance (NMR) and gas/liquid chromatography mass spectrometry (GC/LC-MS) (10-12). The “omics” methods offer an advantage over traditional approaches that is detection of several molecules in a single assay versus investigation of one molecule in each assay.
2. Objectives
The present study was conducted to evaluate whether NMR-based metabolomics may detect patients with NAFLD compared to healthy controls in Iranian population.
3. Methods
3.1. Study Population
Metabolomic analyses were performed on 37 NAFLD patients and on 36 healthy controls. Subjects with NAFLD were enrolled in the study (between January and August 2017) from 22 bahman hospital (Neyshabur, Iran). The NAFLD patients had been newly diagnosed based on hepatic ultrasound. Among 37 NAFLD patients, 8 patients had hyperlipidemia and 3 patients had high blood pressure; therefore, these subjects received medications associated with their disorders. The control group included the volunteers who underwent medical checkups and were verified to have normal liver function. Healthy controls were matched for age and gender with the patient group. To verify the absence of steatosis, all healthy controls also performed hepatic ultrasound.
Blood sample was taken under fasting conditions form each participant in the morning. Serum samples were separated and were stored in aliquots at -80°C until use. The inclusion criteria were: (i) age between 22 - 63 years old; (ii) non-smoker and non-alcoholic people (intake of alcohol for women and men was less than 20 and 30 mg/d, respectively) and exclusion criteria were: (i) having acute or chronic liver disorders including viral and autoimmune hepatitis, alcoholic liver disease and other metabolic liver disorders such as hemochromatosis and Wilson’s disease; (ii) having other medical disorders such as diabetes mellitus, hyper/hypothyroidism, cancer, cardiovascular diseases (iii) being pregnant and lactating women.
The present study protocol was approved by the Ethics Committee of Neyshabur University of Medical Sciences (IR.NUMS.REC.1395.33). A consent form was obtained from all the participants before enrollment in the study.
3.2. Anthropometric and Biochemical Parameters
Body height and weight were measured with an accuracy 0.1 cm and 0.1 kg by a digital stadiometer (Model BSM 370, Seoul, Korea) and InBody (Model 770 device, Seoul, Korea), respectively. Body weight index was calculated by weight/(height)2. All blood specimens were taken in the morning after an overnight fast, then serum samples were separated and biochemical parameters were immediately measured, the rest of serum samples were stored at -80°C until used for metabolomics analyses.
3.3. NMR Acquisition and Data Processing
Preparation of serum for NMR and measurement were performed as explained previously (13, 14). Serum samples were thawed on ice and mixed with 10 % D2O (deuterium oxide, 99.9% D, Aldrich Chemicals Company) for the NMR lock signal, then transferred to 5 mm NMR tubes. NMR analysis was carried out at department of Chemistry at Sharif University of Technology, Tehran, Iran using a Bruker DRX500 MHz spectrometer operating at 500.13 MHz, equipped with 5mm high-quality NMR tubes (Sigma Aldrich., RSA). The 1H NMR analysis were performed at a constant temperature of 298 K and in blinded to the two groups. For all serum samples the Carr-Purcell-Meiboom-Gill (CPMG) spin-echo pulse sequence, π/2 - tD - π - tD experiments were used in order to detect small molecular weight metabolites (15). The 1D spectra acquisition parameters were a 10.5 µs 90° pulse, 128 transients collected into 32 k data points with a spectral width of 8389.26 Hz, an acquisition time of 1.95 seconds and a relaxation delay of 2 seconds. Prior to Fourier transformation, an exponential function corresponding to a line-broadening factor of 0.3 Hz was applied to the free induction decay (FID).
All spectra was referred to solvent and manually phased and baseline corrected by using the XWINNMR software (version 3.5, Bruker Spectrospin Ltd). All spectra were imported to MATLAB (version 6.5.1, The MathWorks, Cambridge, UK) and binned into 408 bins (an equal 0.02 ppm bin width) over a chemical shift in the range between 0.2 and 10 ppm using ProMetab software (version prometab-v3-3) (16) in MATLAB. Serum spectra regions containing water (4.2 - 5.5 ppm) were discarded. H NMR spectra were normalized and aligned by ProMetab in MATLAB. The final data were imported to SIMCA (SIMCA 14.0, Umetrics, Umeå, Sweden) for multivariate statistical analysis.
3.4. Statistical Analysis
3.4.1. The Anthropometric and Clinical Experiments
Comparisons of clinical and anthropometric data were preformed using SPSS 16.0 (SPSS, Inc., Chicago, IL) between control and patient groups. Normality distribution for each variable was evaluated by the Kolmogorov-Smirnov test. Categorical variables between the two groups were compared using Fisher’s exact test. The independent t-test was used for continues variables with normally distributed data.
3.4.2. Metabolomics Experiments
Multivariate statistical analyses were carried out by SIMCA (SIMCA 14.0, Umetrics, Umeå, Sweden). All variables (chemical shifts along with confounding factors) were imported into the SIMCA. First, unsupervised principal component analysis (PCA) was used to identify clusters and outliers in the dataset. Then, supervised orthogonal projections to latent structures-discriminant analysis (OPLS-DA) was applied to recognize differentiation chemical shifts between healthy controls and NAFLD patients. The model quality was described by three parameters (R2X, R2Y and Q2). Moreover, the model prediction performance was evaluated by receiver-operating characteristic (ROC) curve that generated from seven-fold cross validation obtained by SIMCA.
3.5. Metabolite Identification
The metabolites were recognized based on literatures and on-line databases such as human metabolome database (HMDB) (http://hmdb.ca/) and biological magnetic resonance databank (BMRB) (http://bmrb.wisc.edu) according to their chemical shifts obtained by OPLS-DA model.
3.6. Metabolic Pathway Analysis
Metabolite pathway analysis were performed using MetaboAnalyst 3.0. The discriminant metabolites between healthy controls and NAFLD patients were imported into the MetaboAnalyst software (17) in order to seek significant metabolic pathways involved in this disease. The P value and false discovery rate (FDR) < 0.05 were considered statistically significant.
4. Results
4.1. Clinical Patient Characteristics
In this study, we analyzed the metabolic profiles of 75 serum samples collected from 37 healthy controls and 38 patients with NAFLD for NMR analysis (two samples were identified as outliers and deleted from analysis) that their anthropometric and biochemical parameters were summarized in Table 1. Of the 37 participants, 19 were with grade 1 and 18 with grade 2. In the NAFLD and control groups, the gender ratio showed no significant difference. Age and height were similar in the NAFLD and control groups, whereas body weight and BMI were significantly higher in the NAFLD than in the control group. Also, NAFLD patients had significantly higher levels of serum total cholesterol, triglycerides and hepatic enzymes including alkaline phosphatase, aspartate aminotransferase, and alanine aminotransferase compared to the healthy controls (Table 1).
| Control (N = 36) | NAFLD (N = 37) | P Value | |
|---|---|---|---|
| Gender, M/F | 13/23 | 17/20 | 0.478 |
| Age, y | 35.61 ± 1.72 | 38.55 ± 1.46 | 0.199 |
| Height, cm | 164.7 ± 1.46 | 164.45 ± 1.9 | 0.914 |
| Weight, kg | 66.35 ± 1.76 | 79.26 ± 2.07 | 0.000 |
| BMI, kg/m2 | 24.44 ± 0.57 | 29.46 ± 0.81 | 0.000 |
| FBS, mg/dL | 91.05 ± 1.11 | 92.19 ± 2.05 | 0.628 |
| TC, mg/dL | 170.82 ± 5.56 | 194.02 ± 7.29 | 0.014 |
| TG, mg/dL | 109.02 ± 8.49 | 147.83 ± 12.22 | 0.011 |
| LDL-C, mg/dL | 100.91 ± 4.24 | 103.97 ± 4.77 | 0.633 |
| HDL-C, mg/dL | 45.8 ± 1.21 | 45.01 ± 1.68 | 0.708 |
| ALT, IU/L | 21.51 ± 1.79 | 41.72 ± 4.69 | 0.000 |
| AST, IU/L | 18.11 ± 0.98 | 28.77 ± 2.75 | 0.001 |
| ALP, U/L | 160.42 ± 7.14 | 191.33 ± 9.24 | 0.01 |
| T Bili, mg/dL | 0.79 ± 0.04 | 0.83 ± 0.06 | 0.613 |
| D Bili, mg/dL | 0.21 ± 0.01 | 0.24 ± 0.01 | 0.202 |
Abbreviations: ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; D Bili, direct bilirubin; FBS, fasting blood sugar; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; T Bili, total bilirubin; TC, total cholesterol; TG, triglyceride.
aValues are presented as mean ± SE.
bComparison between two groups (control vs. NAFLD patients) using the independent t-test and Fisher's exact test (for categorical variables such as gender).
4.2. Discrimination Between NAFLD and Control Groups Using Multivariate Analysis
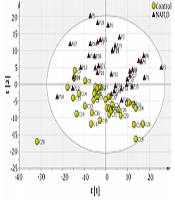
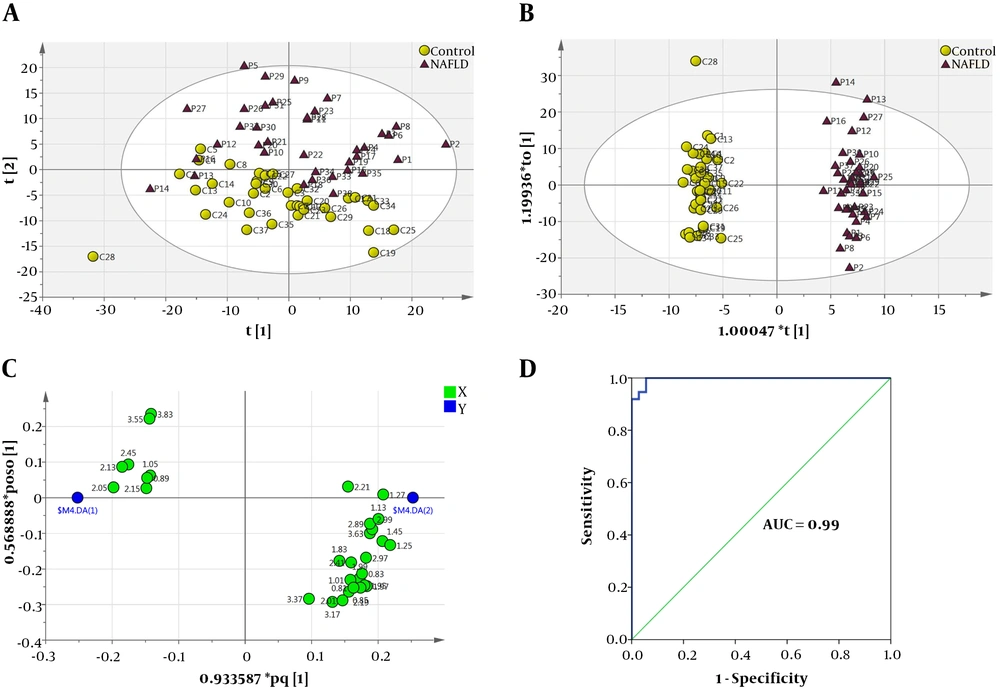
The obtained matrix containing the numbers of samples and 409 variables (408 chemical shifts along with BMI as confounding factor) was imported into SIMCA and analyzed using unsupervised PCA for detecting outliers. The PCA scoter plot showed two serum samples were found to be outside the 99% Hotelling’s T2 confidence limit and were removed for further analysis (Appendix A and B). After removing two outliers, the final PCA was performed (Figure 1A: R2X = 0.857, Q2 = 0.729). Then, the OPLS-DA was performed to identify potential metabolic biomarkers between the two study groups (Figure 1B: R2X = 0.67, R2Y = 0.982, Q2 = 0.924). The loading plot of OPLS-DA model has been illustrated in Figure 1C. This loading plot demonstrated the chemical shifts of the NMR spectra that are responsible for clustering in OPLS-DA score plot. To validate the OPLS-DA model ROC curve was used, the area under the ROC curve (AUC) value was 0.99 for the OPLS-DA model (Figure 1D).
Multivariate statistical analysis from NMR-based metabolic profiling. A, PCA score plot after excluding outliers; B, OPLS-DA score plot of the two study groups (yellow circle, control; purple triangle, NAFLD patients); C, OPLS-DA loading plot demonstrating discriminant variables between the two study groups (the variables on the right are elevated chemical shifts and the variables on the left are reduced chemical shifts in NAFLD patients compared with control group, related metabolites are tabulated in Table 2). D, ROC curve analysis was performed to assess the diagnostic performance of the model, AUC value was 0.99.
4.3. Metabolite Identification and Pathway Analysis of Altered Profiles
Our findings showed nineteen different metabolites that were recognized based on the literatures and on-line databases. Chemical shifts and signal multiplicity were used for identification of metabolites. Compared to healthy controls, NAFLD patients had increased serum concentrations of glycochenodeoxycholic acid, taurocholic acid, glycocholic acid, deoxycholic acid, valine, isoleucine, succinic acid, isocitric acid, 2-ketoglutaric acid, trimethylamine, proline, hydroxyproline and tyrosine, while the concentrations of butyric acid, propionic acid, isovaleric acid, glutamine, glycine, and serine decreased (Table 2).
| Metabolite | δ 1H (p.p.m.)a | Fold Change | Direction of Variationb | Metabolic Pathway |
|---|---|---|---|---|
| 1- Butyric acid | 0.89, 2.13 | 1.9 | ↓ | Gut microbiome-derived metabolism |
| 2- Propionic acid | 1.05, 2.15 | 2.29 | ↓ | Gut microbiome-derived metabolism |
| 3- Isovaleric acid | 2.05 | 2.05 | ↓ | Gut microbiome-derived metabolism |
| 4- Glutamine | 2.45 | 2.36 | ↓ | Amino acid metabolism |
| 5- Glycine | 3.55 | 1.67 | ↓ | Amino acid metabolism |
| 6- Serine | 3.83 | 1.5 | ↓ | Amino acid metabolism |
| 7- Glycochenodeoxycholic acid | 0.83, 2.01, 2.19 | 1.83 | ↑ | Bile acid biosynthesis |
| 8- Taurocholic acid | 0.81, 0.85, 1.95, 3.63 | 2.17 | ↑ | Bile acid biosynthesis |
| 9- Glycocholic acid | 1.25, 1.83 | 4.15 | ↑ | Bile acid biosynthesis |
| 10- Deoxycholic acid | 1.13, 2.21 | 4.46 | ↑ | Bile acid biosynthesis |
| 11- Valine | 1.01 | 1.68 | ↑ | BCAA metabolism |
| 12- Isoleucine | 1.27, 1.45, 1.97 | 2.29 | ↑ | BCAA metabolism |
| 13- Succinic acid | 2.41 | 1.82 | ↑ | TCA cycle |
| 14- Isocitric acid | 2.97 | 1.68 | ↑ | TCA cycle |
| 15- 2-Ketoglutaric acid | 2.99 | 1.67 | ↑ | TCA cycle |
| 16- Trimethylamine | 2.89 | 2.01 | ↑ | Gut microbiome-derived metabolism |
| 17- Proline | 1.99 | 1.8 | ↑ | Amino acid metabolism |
| 18- Hydroxyproline | 3.37 | 1.82 | ↑ | Amino acid metabolism |
| 19- Tyrosine | 3.17 | 2.77 | ↑ | Amino acid metabolism |
Abbreviations: BCAA, branched-chain amino acid; TCA: tricarboxylic acid cycle.
aChemical shift scale of NMR signal used for quantification of metabolites
bIncreased or decreased metabolites in NAFLD group compared with control group.
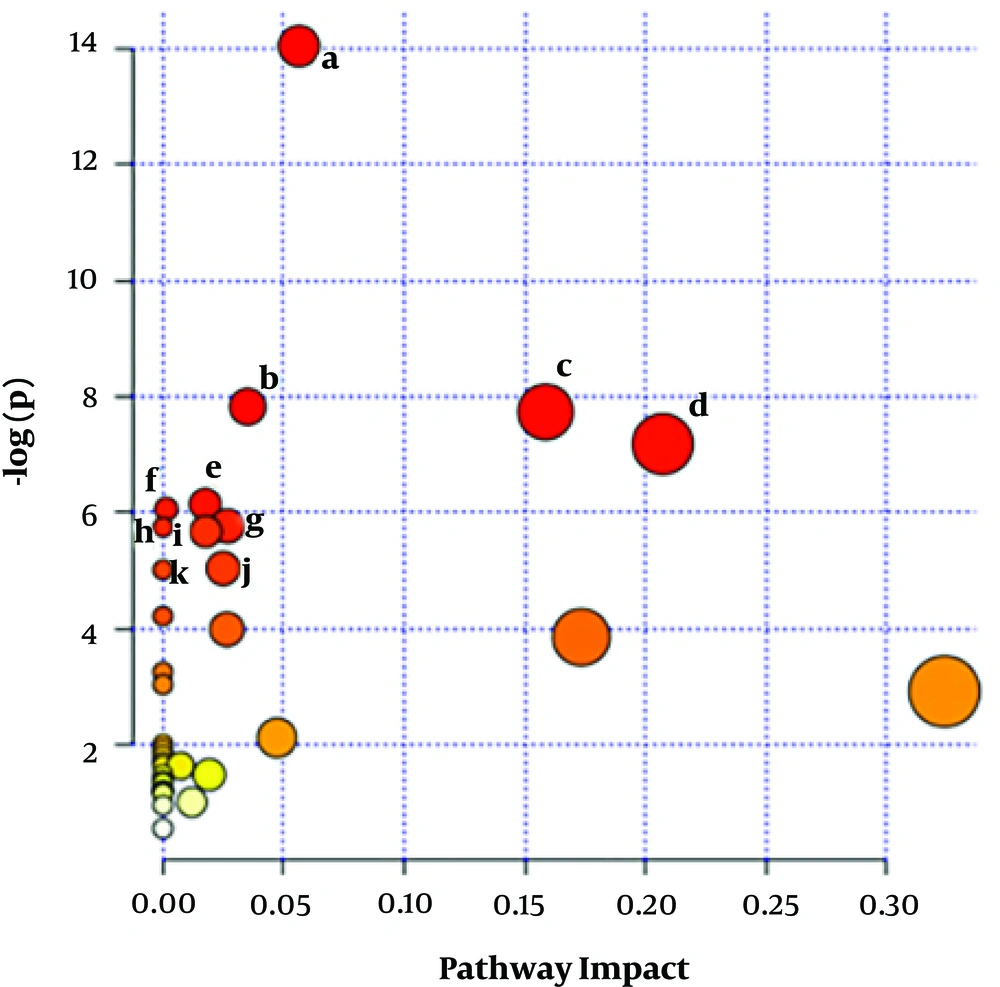
Pathway analysis was performed on metabolites detected via NMR analysis using the MetaboAnalyst 3.0 servers. This software showed eleven metabolic pathways changed in serum of patients with NAFLD. These pathways included aminoacyl-tRNA biosynthesis, primary bile acid biosynthesis, citrate cycle (TCA cycle), alanine, aspartate and glutamate metabolism, methane metabolism, propanoate metabolism, D-glutamine and D-glutamate metabolism, nitrogen metabolism, butanoate metabolism, glyoxylate and dicarboxylate metabolism, cyanoamino acid metabolism (Figure 2 and Table 3).
| Pathway Name | Match Statusa | P Value | FDRb |
|---|---|---|---|
| 1- Aminoacyl-tRNA biosynthesis | 7/75 | 8.0379E-7 | 6.4303E-5 |
| 2- Primary bile acid biosynthesis | 4/47 | 3.9933E-4 | 0.011655 |
| 3- Citrate cycle (TCA cycle) | 3/20 | 4.3707E-4 | 0.011655 |
| 4- Alanine, aspartate and glutamate metabolism | 3/24 | 7.6061E-4 | 0.015212 |
| 5- Methane metabolism | 3/34 | 0.002139 | 0.030465 |
| 6- Propanoate metabolism | 3/35 | 0.0023279 | 0.030465 |
| 7- D-glutamine and D-glutamate metabolism | 2/11 | 0.003113 | 0.030465 |
| 8- Nitrogen metabolism | 3/39 | 0.0031861 | 0.030465 |
| 9- Butanoate metabolism | 3/40 | 0.0034273 | 0.030465 |
| 10- Glyoxylate and dicarboxylate metabolism | 3/50 | 0.0064674 | 0.048247 |
| 11- Cyanoamino acid metabolism | 2/16 | 0.0066339 | 0.048247 |
aThe total number of metabolites in each pathway/the number of identified metabolites in each pathway.
bThe false discovery rate.
Metabolic pathway analysis. MetaboAnalyst 3 displaying changed metabolic pathways in serum from NAFLD group compared with control group. The homosapiens library and the default algorithms were used for enrichment and topology analyses. Statistics for pathways with major change based on the p-value and FDR (P value < 0.05 and FDR < 0.05). These pathways include: a, aminoacyl-tRNA biosynthesis; b, primary bile acid biosynthesis; c, citrate cycle TCA cycle); d, alanine; aspartate and glutamate metabolism; e, methane metabolism; f, propanoate metabolism; g, D-Glutamine and D-glutamate metabolism; h, nitrogen metabolism; i, butanoate metabolism; j, glyoxylate and dicarboxylate metabolism; k, cyanoamino acid metabolism.
5. Discussion
NAFLD is associated with the obesity and alterations in life style affecting many body systems (17). Nevertheless, the pathogenesis is multifactorial and not clearly realized. A few studies have been conducted to find the metabolic biomarkers of this disease. However, because racial and ethnic differences may influence NAFLD prevalence and severity (18, 19), this metabolomic study was performed to detect the metabolic biomarkers and determine the mechanism of progress of NAFLD in Iranian patients.
The present study demonstrated that, 19 metabolites were altered in the patients with NAFLD compared to the control group as discussed below.
5.1. Amino Acid Metabolism
In the present study, a change was observed in the levels of amino acids including, glutamine, aspartate, proline, tyrosine, serine, glycine, isoleucine, leucine, and valine in the patients with NAFLD compared to healthy controls. These findings are not astonishing because most proteins and amino acids are metabolized in the liver. Glutamine, serine and glycine were observed to decrease in the patients with NAFLD compared to healthy controls. These amino acids are as precursors of the most important antioxidants, glutathione (GSH), directly or indirectly (20-22). Our results are in agreement with prior studies that reported reduced levels of these amino acids in plasma of patients with NAFLD (23, 24). However, one study showed that plasma serine did not change significantly in NAFLD patients in comparison with control group (25). Glutamine has been shown to decreases the oxidative stress (OS) through increasing the GSH biosynthesis and ameliorate the hepatic steatosis in the liver of NAFLD rats (26). Besides, Mardinoglu et al. showed that, serine supplementation decreases the hepatic steatosis in humans (27), and another group indicated that, supplementation with glycine might protect against NASH in rats (28).
Our findings indicated high levels of proline and hydroxyproline in the serum of the patients with NAFLD compared to healthy controls. These metabolites are the main components of the collagen protein and are potential biomarkers of elevated hepatic collagen (29). In line with our results, increased serum level of proline has been shown in subjects with NAFLD (30). Also previous studies showed that patients with alcoholic and non-alcoholic liver diseases had high mRNA expression of procollagen compared to the controls (31). Another study showed that, the synthesis of hepatic collagen was elevated in these two groups of patients (32).
Increased levels of tyrosine have been often reported in the serum of patients with NAFLD (23, 33). This is consistent with previous studies, where the aromatic amino acids were associated with the prevalence of liver diseases (34). This finding is not surprising because the liver metabolizes aromatic amino acids, and lipid accumulation in it causes impairment in hepatic metabolism of these amino acids. Down-regulation of aromatic amino acid transporter (SLC16A10) is another possible explanation, as it uptakes these amino acids from blood to liver, in NASH patients (35). Inconsistent with our findings, Mukherjee et al. (25) reported that the levels of plasma tyrosine significantly decreased in patients with NAFLD compared the to control group, but this group did not indicate any explanation for their results.
Our results are in line with previous investigations, which often found that the serum levels of branched chain amino acids (BCAA: isoleucine, leucine, and valine) increased in the subjects with obesity and/or NAFLD (36-38). The previous studies demonstrated that the activity of mitochondrial BCAA-catabolizing enzymes, branched-chain aminotransferase and branched-chain α-ketoacid dehydrogenase (BDKDH), decreased in obese population (39). Because BMI was significantly higher in the patients with NAFLD than the control group; therefore, this cofounding factor was adjusted. Similarly, after adjusting for BMI, an increase in the level of BCAA was shown.
5.2. TCA Cycle
The elevation of circulating BCAA and/or their catabolic intermediate products are involved in the up-regulation of TCA cycle in the liver of these patients (38, 40, 41). As observed, TCA cycle intermediate levels (succinate, α-ketoglutarate and isocitric acid) elevated in the patients with NAFLD compared to the healthy controls in the present study. Moreover, reduced activities of mitochondrial respiratory chain (MRC) complexes demonstrated in this malady can cause OS and mitochondrial dysfunction in the patients with NAFLD (42, 43).
5.3. Gut Microbiome-Derived Metabolites
Dysbiosis, as disruption of the normal gut microflora, has been observed in the NAFLD and metabolic disorders (44). Interestingly, in this study, some metabolites including, short chain fatty acids (SCFAs) (decreased levels of propionic acid, butyric acid, and isovaleric acid), methylamines (increased levels of trimethylamine) and primary and secondary bile acids produced by the fermentation of gut microbiota (44) were observed to alter in the serum of patients with NAFLD compared to the control group.
SCFAs are organic acids with less than 7 carbon atoms and play a main role in attenuating the obesity and dyslipidemia (45, 46). Ruminococcaceae is one of the gut bacteria physiological function of which is producing the SCFAs (47). Previous reports demonstrated a lower proportion of Ruminococcaceae in the patients with NASH compared to healthy subjects (48). Christensenellaceae, another family associated to low BMI in humans were positively correlated with total SCFAs (49). Decreased levels of SCFAs might be because of significantly higher BMI in the patients with NAFLD than the control group. However, after adjusting for BMI, a decrease in the level of SCFAs was also shown.
In the present study, a significant increase was found in the level of primary (glycocholic acid, taurocholic acid, glycochenodeoxycholate) and secondary bile acids (deoxycholic acid (DCA)) in the serum of the patients with NAFLD compared to the control group. Our results are in line with the previous investigations, which showed a higher level of bile acids (BAs) in subjects with hyperlipidemia and NAFLD (24, 50, 51), resulting from (a) increases in synthesis of BAs; (b) defective BA transports. Previous studies indicated that the mRNA expression of farnesoid X receptor (FXR) was down-regulated in patients with NAFLD compared to the controls (52). This protein suppresses cholesterol 7 alpha-hydroxylase (CYP7A1) expression. An increase in the hepatic expression of CYP7A1, the rate-limiting enzyme synthesizing BAs from cholesterol, results in the elevated synthesis of primary bile acids.
As bile acids from hepatic-portal circulation are removed by the liver; therefore, triglycerides accumulation in the liver may cause liver dysfunction and increased levels of bile acids in the circulation in NAFLD patients. Given that NAFLD patients in this study are obese, thus, it can be said that, the obesity causes defective hepatic BA transport and BMI is negatively related to the expression of liver BA transporters (53). Dysbiosis may be another reason for decreased level of secondary bile acids in these patients, as an imbalance in gut flora. Primary BAs are converted into secondary ones through deconjugation and dehydroxylation by intestinal flora. Alterations in gut microbiota have been observed in both NAFLD and obesity (54, 55). For instance, Clostridium cluster XI and XIVa producing DCA, a hydrophobic secondary BA which generates ROS and causes DNA damage, from primary bile acids have been reported to elevate in the obese mice (55).
The present study had some limitations. The diet of two study groups was not evaluated, and also the metabolites were identified by one method (NMR), thus it is recommended to verify our results other approaches.
5.4. Conclusions
The present study was the first investigation assessed the global metabolomics in the Iranian NAFLD patients using the NMR. A total of 19 significantly altered metabolites were detected in the patients compared to the controls. It was found that, some amino acids and their derivatives, bile acids, SCFAs, and TCA cycle intermediates were significantly changed. In brief, our findings demonstrated that, a combination of NMR-based metabolomics along with chemometric techniques offers an effective method for detection of markers of NAFLD with a high accuracy. Our results showed that, precursors of glutathione, glutamine, serine and glycine reduced in NAFLD patients. Therefore, supplementation with these amino acids might be useful for treatment of NAFLD patients. Furthermore, decreased levels of SCFAs could be because of altered levels of gut microbiota, as a result; supplementation with probiotics is another recommendation for treatment of these patients.