1. Context
Chronic viral hepatitis (CVH) is a chronic inflammation of the liver. Several types of human viral hepatitis cause infection either through enteric-transmission (hepatitis A and E) or blood transfusion (hepatitis B, C, and D). Hepatitis A and E are host-controlled infections that do not progress to chronic infection (except for hepatitis E), while hepatitis B, C, and D often progress to persistent chronic infection (1). Hence, host immunity (innate and adaptive) is responsible for the development and progression of diseases. An estimated 257 and 71 million people are living with chronic hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, respectively (WHO 2015 - 2017) (2), making CVH the leading cause for hepatocellular carcinoma (HCC) (60% - 70%) globally, with a total incidence of 16/100000 (3).
Human mucosal-associated invariant T (MAIT) cells were originally identified in 1993, with semi-invariant TCR TRAV1-2/TRAJ33 expression (4). Treiner et al. observed that the canonical hVα7.2-Jα33 or mVα19-Jα33 TCR rearrangement was preferentially located in the gut lamina propria of humans and therefore, named these cells MAIT cells (5). MAIT cells constitute a subpopulation of T cells, including intrahepatic (10% - 40%) and peripheral blood CD 3+T cells (0.1% - 10%) (6, 7). Moreover, they develop in the thymus and migrate to the periphery to become the antigen-specific αβ T-cell population in the human immune system (8). Koay et al. reported PLZF as a controlling transcription factor for three developmental stages of maturation and function of MAIT cells (9). Most studies have defined MAIT cells as CD3+CD161high Vα7.2+ lymphocytes (10-13). MAIT cells recognize Vitamin B2 (riboflavin) metabolites produced by bacteria and yeasts in a major histocompatibility complex-related protein 1 (MR1)-dependent manner (14, 15) and can be activated in an MR1-independent fashion in response to cytokines (7, 16-18). Additionally, MR1 presenting antigen is recognized by MAIT cells using their TCRs (19). Upon TCR-dependent or TCR-independent activation, MAIT cells produce pro-inflammatory cytokines (IFN-γ, TNF-α), cytolytic products (perforin, granulysin and granzymes) and degranulate (exposing CD107a to the cell surface) (20).
MAIT cells have been widely studied in infectious diseases. It is one of the key immune controllers of gut microbiota, fungal infection, bacterial infection, and inflammatory diseases (21, 22). A recently published study showed a lower frequency of MAIT cells, with increased expression of immune exhaustion and chronic immune activation in chronic HCV-infected patients (23). Yong et al. reported the frequency of MAIT cells were significantly reduced (12), while Boeijen et al. reported that MAIT cells were not deleted or functionally impaired in chronic hepatitis B (CHB) patients (24).
2. Objectives
Considering the above reports, we aimed to perform a systematic review and meta-analysis to explore the frequency, phenotype, and function of MAIT cells in patients with CVH.
3. Evidence Acquisition
Our meta-analysis is reported in accordance with the PRISMA (preferred reporting items for systematic reviews and meta-analyses) statement.
3.1. Literature Search
The following electronic databases were used: PubMed, Embase, the Cochrane Library, Google Scholar, ScienceDirect, Web of Science, and the China National Knowledge Infrastructure (CNKI) for all appropriate articles, which published before 25 June 2019. The search strategy and the search terms were developed with the support of two independent reviewers (Qinling Liu and Feng Zhu). The used MeSH (Medical Subject Headings) terms and text words were: “mucosal associated invariant T cells”, “MAIT cells”, “chronic viral hepatitis”, “chronic hepatitis b”, “chronic hepatitis c”, and “chronic hepatitis d”. The language of the enrolled studies was restricted to English or Chinese.
3.2. Selection of Articles
The inclusion criteria were as follows: (a) treatment-free patients with CVH (i.e., patients did not take antiviral therapy or immunosuppressive drugs 6 months before the sampling); (b) the study design: case-control or cohort; (c) study details: frequency, phenotypes or functions of MAIT cells in the peripheral blood or liver tissue.
The following studies were excluded: (a) study of patients co-infected with human immunodeficiency virus; (b) study did not involve healthy controls; (c) duplication or overlapping study; (d) book chapter, review articles or meta-analysis. Our selection decisions were not influenced by the names of the authors or journals of the articles. Third investigator (Dazhi Zhang) was invited to provide arbitration when there was a divergence between two reviewers.
3.3. Data Extraction
The two reviewers scanned the titles and abstracts of all search results independently, and those satisfied the inclusion criteria were selected for full-text review. The final inclusion or exclusion decisions were made by inspecting the full manuscripts. The following data were extracted from each paper: first author and published year, country, type of study design, number of patients and healthy control, characteristics of patients and healthy control, and the results measured by flow cytometry (including frequency, phenotype, and function of MAIT cells). Emails were sent to the corresponding, or co-corresponding authors if the information of the studies were unclear. In addition, if there were no email replies from the authors, the software GetData Graph Digitizer 2.22 was used to get data from the articles.
3.4. Quality Assessment and Risk of Bias
The Newcastle-Ottawa Scale (NOS) was applied for assessing the quality of studies (Wells et al., 2014) (25). The score of NOS ranged from zero to nine points, and the assessment of study quality is shown in Table 1. The publication bias of the meta-analysis was evaluated with Begg’s and Egger’s tests using STATA version 15.1
| Study | Study Type | Selection | Comparability | Exposure/Outcome | Total |
|---|---|---|---|---|---|
| Provine et al. (26), 2018 | Case-control | *** | * | ** | ****** |
| Yong et al. (27), 2018 | Case-control | *** | ** | ** | ******* |
| Setsu et al. (28), 2017 | Case-control | ** | * | ** | ***** |
| Boeijen et al. (24), 2017 | Case-control | ** | ** | ** | ****** |
| Beudeker et al. (29), 2018 | Case-control | ** | ** | ** | ****** |
| Bolte et al. (30), 2017 | Cohort | *** | * | *** | ****** |
| Dias (31), 2019 | Case-control | ** | ** | ** | ****** |
| Spaan et al. (32),2016 | Cohort | *** | ** | *** | ******** |
| van Wilgenburg et al. (33), 2016 | Case-control | *** | * | ** | ****** |
| Barathan et al. (23), 2015 | Case-control | *** | ** | ** | ******* |
3.5. Statistical Analysis
After collecting and converting, data of the included studies were combined by meta-analyses using Review Manager 5.3 (The Nordic Cochrane Centre, The Cochrane Collaboration, 2012). I2 > 50% indicated there was significant heterogeneity then random effects model was applied, otherwise affixed effects model was applied (34). Standard mean difference (SMD) with a 95% confidence interval (CI) was used to analyze continuous variables, including frequency, phenotype, and function of MAIT cells. A P value of < 0.05 was considered statistically significant.
4. Results
4.1. Search Results and Study Characteristics
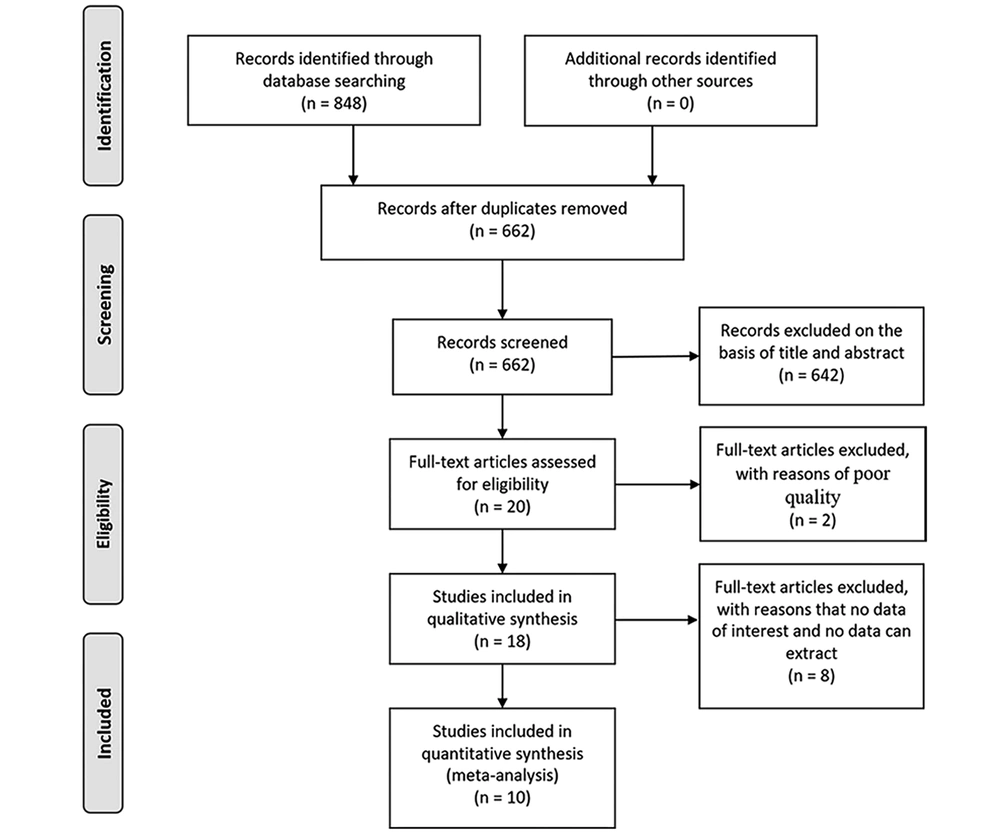
Flow diagram of study selection is shown in Figure 1. A total of 848 studies were identified adopting the search strategy. Of these twenty-one publications were from PubMed, 57 from Embase, 46 from Web of Science, 2 from Cochrane Library, 620 from Google scholar, 101 from ScienceDirect and 1 publication was identified from CNKI. After removing 186 duplicates and excluding 226 review, 53 book chapter, and 363 irrelevant records, 20 full-text were assessed for eligibility. One letter and one abstract of congresses were removed because of poor quality. Four full-text articles were excluded because of having no data of interest and four studies were excluded because of no way to extract data. Finally, ten studies were included in the meta-analysis (23, 24, 26-33).
The characteristics of the included studies are listed in Table 2. Among the final inclusion, eight studies were case-control studies and two were cohort studies. Three studies were from the Netherlands, two studies were from the United Kingdom, two from Malaysia, one from Japan, one from the USA, and one from Sweden.
| Study, Patients No. | Age in Years | Male, % | ALT (U/I) | HBV-DNA or HCV-RNA |
|---|---|---|---|---|
| Provine et al. (26), 2018, United Kingdom | ||||
| Healthy controls: 16 | NA | NA | NA | - |
| Chronic HCV: 33 | 45 (20 - 76)a | 54.55 | 59 (21 - 206)a | NA |
| Yong et al. (27), 2018, Malaysia | ||||
| Healthy controls: 13 | 44 (40 - 51)b | 61.5 | NA | - |
| HBV-DNA + ve: 12 | 50 (47.3 - 57.3)b | 50 | 26.5 (18.3 - 40.3)b | 20377 (3468 - 326353)b, g |
| HBV-DNA - ve: 9 | 55 (38 - 61)b | 66.7 | 45 (20 - 36.5)b | - |
| Setsu et al. (28), 2017, Japan | ||||
| Healthy controls: 13 | 66 (44 - 83)a | 57.89 | 47 (11 - 209)a | - |
| Chronic viral hepatitis: 19 | 52 (30 - 81)a | 61.54 | 14 (9 - 26)a | NA |
| Boeijen et al. (24), 2017, The Netherlands | ||||
| Healthy controls: 17 | 41h | 64.71 | NA | - |
| Chronic hepatitis B: 18 | 43 (25 - 59)c | 72.22 | 58 (14 - 366)c | 7.61 (2.46 - 8.57)c, f |
| Beudeker et al. (29), 2018, The Netherlands | ||||
| Healthy controls: 9 | 40 (28 - 50)e | 100 | NA | - |
| HCV F0- F1: 9 | 54 (45 - 68)e | 100 | 57 (24 - 95)e | 4.38 × 106e, f |
| HCV F3- F4: 11 | 51 (47 - 57)e | 100 | 100(38 - 211)e | 2.90 × 106e, f |
| Bolte et al. (30), 2017, USA | ||||
| Healthy controls: 42 | NA | NA | NA | - |
| Chronic HCV: 56 | 59 (55.0 - 62.3)b | 57 | 67 (36 - 128)b | 6.4 (5.6 - 5.9)b, f |
| Dias et al. (31), 2019, Sweden | ||||
| Healthy controls: 57 | 42 (21 - 65)a | 46 | NA | - |
| Chronic hepatitis B: 38 | 42 (26 - 74)a | 28 | 26 (9 - 98)a | 341 (5 - 33000000)a, g |
| Chronic HDV: 41 | 38 (19 - 61)a | 51 | 76 (31 - 360)a | HDV: 94190 (0 - 23000000)a, g; HBV: 152 (0 - 1.1E + 09)a, h |
| Spaan et al. (32),2016 The, Netherlands | ||||
| Healthy controls: 22 | 54 (42 - 70)c | 67 | NA | - |
| CHCV PegIFN/riba: 11 | 45 (27 - 60)c | 73 | 79 (34 - 164)c | 6.66 (2.57 - 7.43)c, f |
| CHCV PegIFN/riba/telaprevir: 11 | 50 (25 - 61)c | 82 | 70 (29 - 140)c | 6.41 (4.54 - 6.91)c, f |
| CHCV Asunaprevir/daclatasvir: 5 | 52 (43 - 66)c | 80 | 188 (94 - 269)c | 6.08 (5.2 - 6.36)c, f |
| CHCV Sofosbuvir/daclatasvir: 6 | 59 (36 - 70)c | 67 | 126 (24 - 196)c | 6.43 (4.92 - 6.72)c, f |
| van Wilgenburg et al. (33), 2016, United Kingdom | ||||
| Healthy controls:23 | NA | NA | NA | - |
| Chronic HCV: 17 | 51 (32 - 68)c | 58.82 | NA | NA |
| Barathan et al. (23), 2015, Malaysia | ||||
| Healthy controls: 25 | 26 (24 - 33)c | NA | NA | - |
| Chronic HCV: 25 | 40 (29 - 55)c | 60 | NA | 151613 (15 - 1310000)c, g |
aMedian (range).
bMedian (IQR).
cMean (range).
dMean.
eUnspecified mean or median.
fLog10IU/mL.
gCopies/mL.
hIU/mL.
4.2. Frequency of Peripheral MAIT Cells
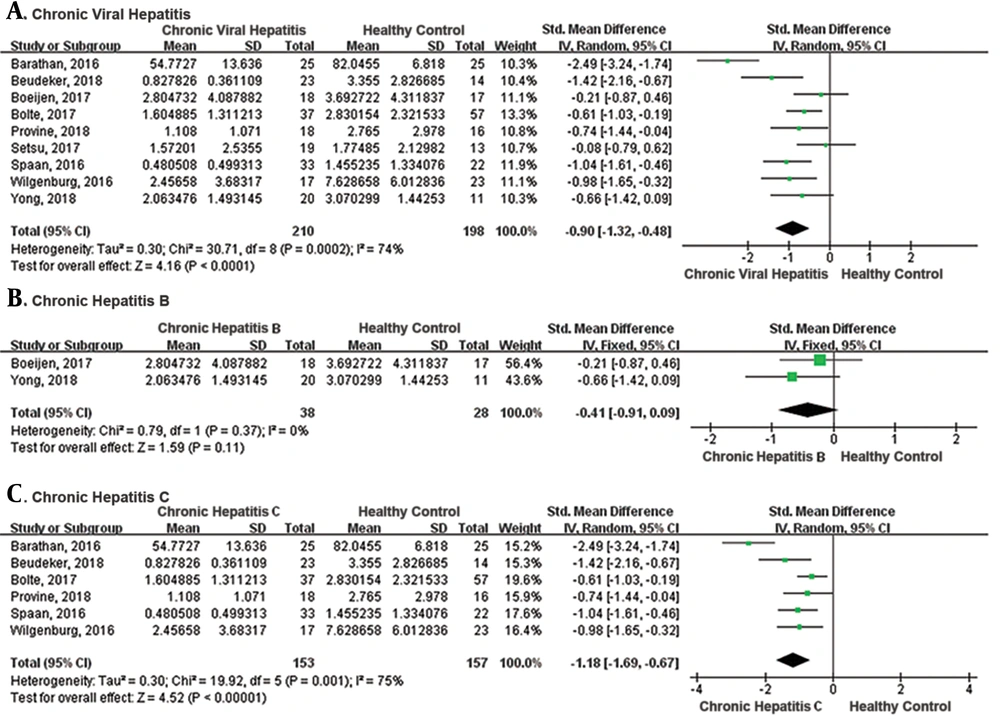
The meta-analysis of the 9 studies showed that there was a significantly lower frequency of peripheral MAIT cells in CVH patients compared with healthy control (SMD = -0.90, 95% CI: -1.32 to -0.48, P < 0.0001; Figure 2). The I2 value was 74% indicating significant heterogeneity and random effect model was applied. One study identified MAIT cells by different gating orders, which may result in significant heterogeneity. There was no significant difference in the frequency of MAIT cells in chronic hepatitis B subgroup compared with healthy controls (SMD = -0.41, 95% CI: -0.91 to 0.09, P = 0.11). The I2 value was 0% indicating no significant heterogeneity and fixed effects model was applied. The frequency of MAIT cells was significantly lower in chronic hepatitis C subgroup than healthy control (SMD = -1.18, 95% CI: -1.69 to -0.67, P < 0.00001). The I2 value was 75%, indicating significant heterogeneity and random effects model was applied.
4.3. Phenotype of Peripheral MAIT Cells
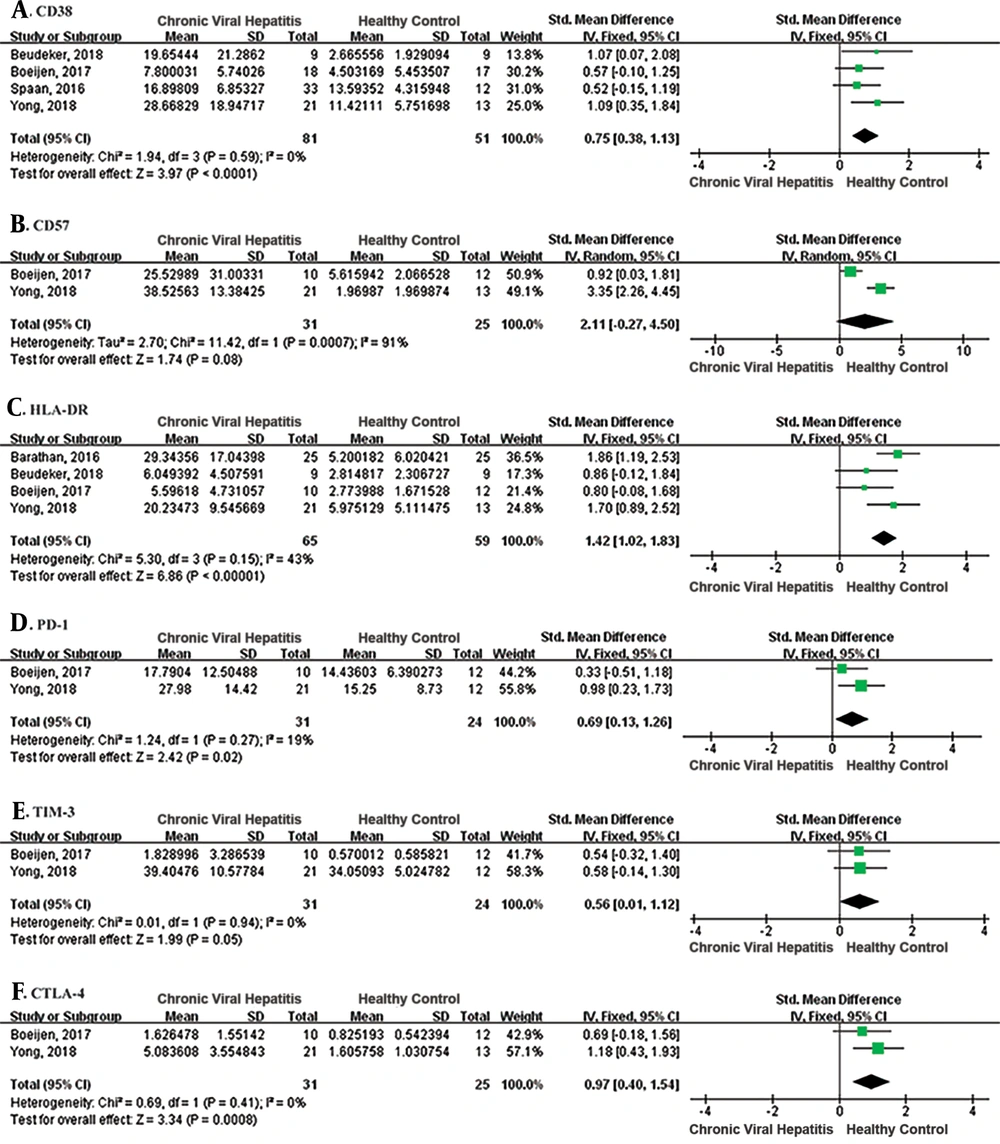
To investigate the phenotype of MAIT cells, the expression of HLA-DR, CD38, CD57, PD-1, TIM-3, and CTLA-4 of MAIT cells in the blood of patients with CVH and healthy controls were analyzed (Figure 3). The pooled SMD of the CD38 expression showed there was a significantly higher level of MAIT cells in patients with CVH than healthy controls (SMD = 0.75, 95% CI: 0.38 to 1.13, P < 0.0001). The I2 value was 0%, indicating no statistical heterogeneity and the fixed effects model was applied. A meta-analysis of the four studies that evaluated HLA-DR also showed MAIT cell levels were significantly higher in patients with CVH than healthy controls (SMD = 1.42, 95% CI: 1.02 to 1.83, P < 0.00001). The I2 value was 43%, indicating no statistical heterogeneity and the fixed effects model was applied. Meta-analysis of CD57 showed no significant difference between the two groups (SMD = 2.11, 95% CI: -0.27 to 4.50, P = 0.08). Meta-analysis of two studies that evaluated PD-1 and CTLA-4 showed a significant difference between patients with CVH and healthy controls (PD-1: SMD = 0.69, 95% CI: 0.13 to 1.26, P = 0.02; CTLA-4: SMD = 0.97, 95% CI: 0.40 to 1.54, P = 0.0008). However, there was no significant difference in the expression of TIM-3 (SMD = 0.56, 95% CI: 0.01 to 1.12, P = 0.05). In conclusion, MAIT cells displayed an activated and exhausted phenotype during CVH.
4.4. Function of Peripheral MAIT Cells
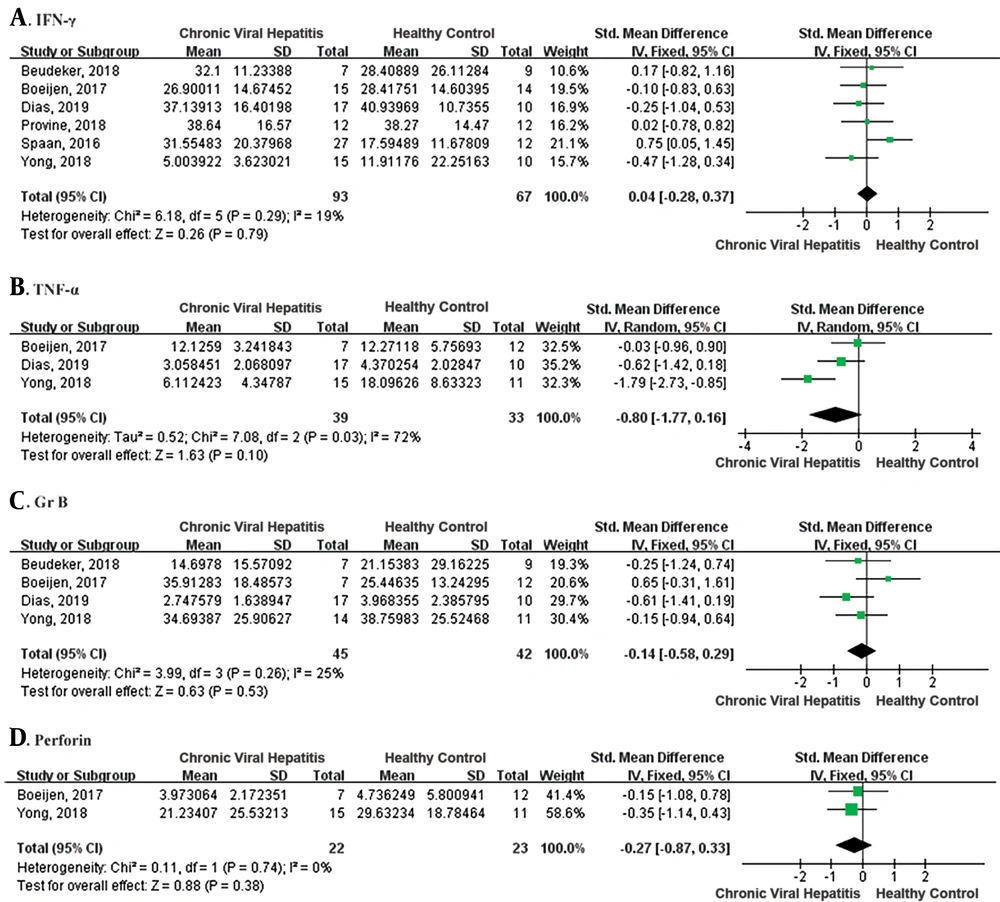
MAIT cells respond to antigen (TCR-dependent) and cytokine (TCR-independent) stimulation by producing proinflammatory cytokines, including IFN-γ, TNF-α, and IL-17, and exert cytotoxic effector functions by releasing granzyme B and perforin (6). We evaluated IFN-γ, TNF-α, perforin, and granzyme B production of peripheral MAIT cells in patients with CVH and the healthy controls (Figure 4). There was no difference in IFN-γ and TNF-α production with MAIT cells in patients with CVH and healthy controls (IFN-γ: SMD = 0.04, 95% CI: - 0.28 to 0.37, P = 0.79; TNF-α: SMD = -0.80, 95% CI: -1.77 to 0.16, P = 0.10). I² were 19% and 72%, and fixed effects model and random effects model were applied, respectively. Additionally, there was also no difference in perforin and granzyme B releasing (perforin: SMD = -0.27, 95% CI: -0.87 to 0.33, P = 0.38; granzyme B: SMD = - 0.14, 95% CI: -0.58 to 0.29, P = 0.53). I2 were 0% and 25%, indicating no statistical heterogeneity and fixed effects model was applied. Therefore, the meta-analysis of the function of MAIT cells showed no significant difference between patients with CVH and healthy controls.
4.5. Publication Bias
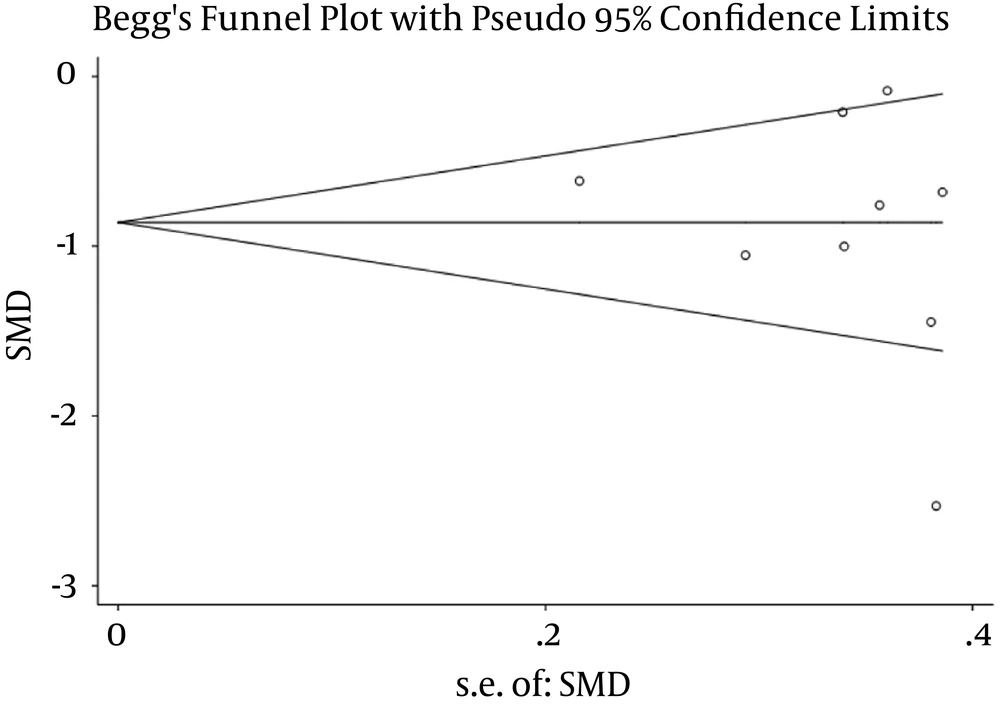
The publication bias of the meta-analysis was assessed by Begg’s tests and Egger’s tests. The result showed no significant publication bias in this study (Figure 5).
5. Conclusions
Our study is the first attempt to review the original study in order to provide a comprehensive estimate of abnormal immune profiles of MAIT cells in CVH. MAIT cells in patients with CVH displayed an activated and exhausted phenotype, which might lead to the reduction of MAIT cells in peripheral blood; nevertheless, no impairment was seen in MAIT cells’ function compared to healthy controls.
Given the essential role of MAIT cells in infectious diseases and inconsistent results of available studies, we performed a meta-analysis to investigate the immune profiles of MAIT cells in CVH. Our study shows that the frequency of peripheral MAIT cells in patients with CVH was significantly reduced compared to healthy controls. In line with our findings, previous studies have also shown the level of peripheral MAIT cells were significantly lower in infectious diseases such as HIV infection (35) and primary biliary cholangitis (10). MAIT cells have been reported to home to peripheral tissues, especially the intestine and the liver with high expression of chemokine receptors CCR5, CCR6, and CXCR6 (36). Previous studies showed that the disappearance of MAIT cells from the peripheral blood was associated with modified expression of chemokine receptors (10, 23). MAIT cells are also identified as IL-18R+Vα7.2+ T-cell population, significantly losing their CD161 expression in HIV-infected patients compared with uninfected controls (11). This suggests that increasing MAIT cells are recruited to inflammatory sites and down-regulation of CD161 may lead to the reduction of peripheral blood MAIT cells in patients with CVH.
In agreement with current studies, MAIT cells display an activated and exhausted phenotype in patients with CVH, with higher levels of immune activation markers (CD38 and HLA-DR) and inhibitory markers (PD-1 and CTLA-4). MAIT cells are unable to recognize viruses in TCR-dependent way due to insufficient production of Vitamin B2 (riboflavin) metabolites. However, viruses can activate MAIT cells via stimulating cytokine production. Some researchers also point out that viruses activate MAIT cells by stimulating cytokine release via ligation of toll-like-receptors (TLRs) or other pattern recognition receptors (PRRs) (37). TLR8 agonist, ssRNA40, can indirectly activate MAIT cells dependent on IL-12 and IL-18 production intrahepatic monocytes (38). Overactivation of MAIT cells may also progress to apoptosis, leading to the reduction of MAIT cells.
Contrary to our expectation, the function of MAIT cells was not impaired in patients with CVH compared to healthy controls. Co-culturing MAIT cells with non-typeable Haemophilus influenzae-infected macrophages (18) or Francisella novicida-infected macrophages (39) can activate MAIT cells. A study also showed that DENV, influenza virus, and HCV-exposed dendritic cells (DCs) or macrophages could activate MAIT cells (33). Moreover, stimulating MAIT cells with anti-CD3/CD28 double beads (28, 40), mitogenic phorbol myristate acetate (PMA)/Ionomycin (24, 27, 41-43), E. coli (24, 30, 44) or cytokines such as IL-12 and IL-18 (12, 26, 30, 42, 44), are usually used in function assay. In our five included studies for function assay, three articles applied IL-12/IL-18 stimulation, one applied double beads, and one applied IL-12/IL-18/E.coli, which may partly influent the results of the meta-analysis.
There are several limitations to this study. First, only a few studies were included, and the size of the included studies was small. Second, the authors’ silences to email replies prevented us from collecting original data; as a result, the software GetData Graph Digitizer 2.22 was used to get data from the articles. Third, there was not enough study to analyze the intrahepatic MAIT cells and compare MAIT cells before and after the treatment. Therefore, future studies should be conducted to provide insight into the mechanisms of CVH to yield a new and better treatment strategy against CVH.
5.1. Conclusions
Our meta-analysis demonstrates a significant depletion of MAIT cells in the peripheral blood of patients with CVH (hepatitis B and C subgroups) compared to healthy controls. In addition, MAIT cells in patients with CVH displayed an activated and exhausted phenotype, which might result in the reduction of MAIT cells; however, no impairment was observed in cytolytic function and cytokine production of these cells.