1. Introduction
Acute-on-chronic liver failure (ACLF) is a syndrome characterized by acute decompensation of chronic liver disease, and is associated with high short-term mortality. Severe infection is a major risk factor for mortality in patients with liver failure (1).
Carbapenem-resistant Enterobacteriaceae (CRE) infection, associating with high mortality, has become a serious health problem worldwide. As one of the most frequently encountered CREs in healthcare setting, carbapenem-resistant Klebsiella pneumoniae may result in fatal infection with limited antibiotic options. It is more worrisome that Klebsiella pneumoniae carbapenemases (KPCs) are highly transmissible and have emerged as a global threat (2). The global spread of multidrug-resistance (MDR) plasmids has been enhanced by selective pressure from antibiotic usage in human and veterinary medicine (2). Owing to limited treatment, high mortality highlights the necessity of exploring additional therapeutic approaches.
To date, knowledge about treatment of the multi-drug resistant K. pneumoniae infection in patients with severe hepatitis or liver failure is limited. Herein, we attempted to report the combination of meropenem with ertapenem as salvage therapy for a patient with ACLF and KPC-producing K. pneumoniae infection.
2. Case Presentation
A 70-year-old Chinese woman, who was diagnosed with ACLF (1) and decompensated liver cirrhosis, was admitted to our department on 9 November 2018. She underwent a transjugular intrahepatic portosystemic stent shunt (TIPSS) one year ago, and consumed warfarin tablet daily (2.5 mg). Two weeks before her admission in our department, she had spontaneous intracerebral hemorrhage, and then warfarin was withdrawn. After drainage of intracranial hematoma, she stayed in intensive care unit (ICU) for 12 days, and was then transferred to our department because of the liver failure. The patient’s clinical data were obtained during 8 months of follow-up, and the patient signed a written informed consent form as well. This study was approved by the Ethics Committee of the Third People’s Hospital of Changzhou (Changzhou, China), and conducted in accordance with the 1975 Declaration of Helsinki.
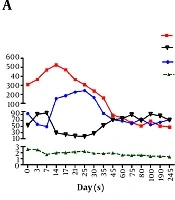
The serum total bilirubin (TBIL) level was 308.7 µmol/L at the time of admission, and the international normalized ratio (INR) was 2.56. Entecavir was given due to hepatitis B virus (HBV) DNA-positive (3.5 E+05 IU/mL, Cobas Taqman HBV test, Roche AG, Basel, Switzerland) at the time of admission. The serum level of TBIL ascended to 526 µmol/L on the 14th day after admission, and then gradually declined (Figure 1A). The INR was maintained above 1.5 for 2 months. In addition, creatinine level significantly increased (from 92.5 to 241 µmol/L) in the first month after admission, while estimated glomerular filtration rate (eGFR) declined to 16.9 mL/min 1.73m2. Diammonium glycyrrhizinate, plasma, as well as albumin were given intravenously.
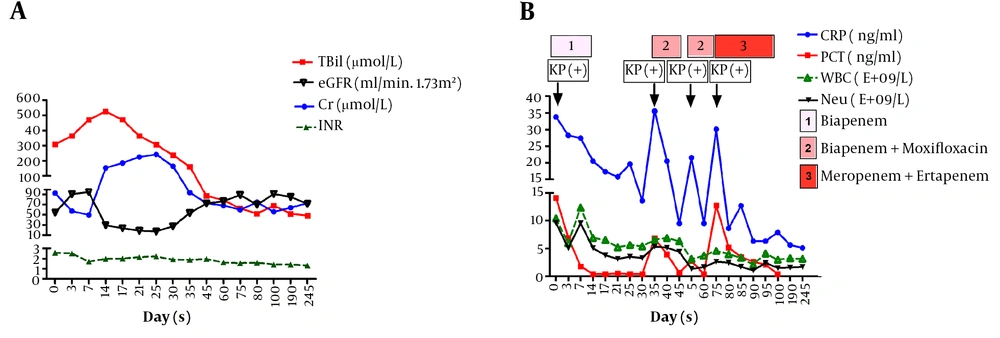
At the time of admission, she had sepsis with symptoms, including fever and shivering, and K. pneumoniae was identified by blood culture test. Biochemical tests showed high levels of procalcitonin (PCT, 14.1 ng/mL) and C-reactive protein (CRP, 33.7 mg/L). Biapenem (0.3 g, q12h) was administered at the time of admission (Figure 1B). Then, antimicrobial susceptibility testing using VITEK-2 system (bioMerieux, Marcy-l'Étoile, France) showed that K. pneumoniae isolate was multidrug resistant according to the minimal inhibitory concentrations (MICs, μg/mL) of antibiotics, involving ceftazidime (≥ 64), ceftriaxone (≥ 64), cefepime (≥ 64), cefazolin (≥ 64), cefotetan (≥ 64), cefoperazone-sulbactam (≥ 64), ampicillin (≥ 32), ampicillin-sulbactam (≥ 32), piperacillin-sulbactam (≥ 128), aztreonam (≥ 64), amikacin (≥ 64), macrodantin (≥ 512), meropenem (≥ 16), imipenem (≥ 16), ertapenem (≥ 8), ciprofloxacin (≥ 4) , levofloxacin (≥ 8), tobramycin (≥ 16), Fosfomycin (≥ 16), and tigecycline (≥ 16).
However, clinical manifestations were improved on the second day after biapenem administration. On the 14th day, biapenem was withdrawn according to the normal temperature and PCT. On the 35th day, fever and shivering re-occurred, and K. pneumoniae became positive in blood culture test. Repeated drug susceptibility tests confirmed the same multidrug-resistant K. pneumoniae isolate without extended-spectrum β-lactamases (ESBLs). Biapenem (0.3 g, q12h) combined with moxifloxacin (0.4 g, qd) was shown to be effective, while that couldn’t eliminate the bacteria. The symptoms of sepsis appeared several times through a positive blood culture, and then, combination of meropenem (2 g, q8h) and ertapenem (1 g, q24h) was administered on 22 January 2019. Furthermore, a negative blood culture result was noted, and the symptoms of sepsis disappeared 3 days after combination therapy. Besides, 28 days after combination therapy, a successful treatment outcome was achieved. Three months after admission, the patient recovered without liver transplantation or artificial liver supporting treatment, and she was discharged on 19 February 2019.
Whole genome sequencing of the multidrug resistant K. pneumoniae isolate was performed by Novogene co. LTD (Beijing, China) using single-molecule real-time (SMRT) sequencing. In brief, the reads were filtered by SMRT Link V. 5.0.1 (Pacific Biosciences of California, Inc., Menlo Park, CA, USA), and assembled to generate a single contig without any gaps. The isolate was identified as V113 according to the 7 core genes, including gapA, infB, mdh, pgi, phoE, rpoB, and tonB (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html). In the present V113 isolate, 3 genes (blaPKC, blaTXM, and catA2) on the plasmid and 3 genes (blaSHV2, catB3, and arnA) on the chromosome were identified. Moreover, 10 multidrug resistance efflux pump genes and 2 fosmidomycin efflux pump genes were found on the chromosome. Moreover, comparative genomic analysis of K. pneumoniae with other Klebsiella strains may assist scholars to find out the evolution of the isolate (supplementary file Appendix 1). Phylogenetic tree of K. pneumoniae based on whole genome sequencing showed that the core genome of K. pneumoniae was similar to that of HS11286.
3. Discussion
In the present study, a 70-year-old patient with ACLF and multidrug-resistant K. peumoniae isolate infection was described. Prolonged treatment with meropenem and ertapenem resulted in a successful treatment outcome without deterioration of liver function. The genome of multidrug-resistant K. pneumoniae was analyzed using SMRT sequencing.
In the present study, HBV infection was found as the leading cause of liver failure, and entecavir was given due to being HBV DNA-positive at the time of patient’s admission. K. pneumoniae infection may perform as a risk factor for the aggravation. Knowledge about multidrug-resistant K. pneumoniae infection in patients with liver failure was scarce. The proportion of KPC-producing K. pneumoniae strains has remarkably increased year-by-year, associating with high mortality (3, 4). Recently, Athans et al. (5) reported that persistent KPC-producing K. pneumoniae infection could be resolved after initiating meropenem-vaborbactam. Colistin, which has a narrow therapeutic window, has been previously recommended, however, it has still remained controversial in patients with renal dysfunction (5). Double-carbapenem regimen (meropenem and ertapenem) was reported to be an effective option to treat KPC-producing CRE infection, even in patients with meropenem MICs (6, 7). Ertapenem which has stronger bonding capability to KPC, is degraded preferentially, and then, the other kinds of carbapenems can be protected (6). To date, the combination therapy of meropenem and ertapenem in patients with liver failure has not been reported. Additionally, in the current study, it was revealed that the mentioned combination therapy is effective without an obvious toxicity. These results suggest that combination of meropenem with ertapenem may serve as an alternative therapeutic strategy for KPC-producing K. pneumoniae infection, even in patients with ACLF.
To date, a small number of studies have assessed the molecular characteristics of multidrug-resistant K. pneumoniaeST147, H11, MGH78578 isolates using single molecule real-time (SMRT) sequencing (8-10). However, multidrug-resistant K. pneumoniae V113 isolate has not been reported, especially in patients with severe hepatitis. Furthermore, SMRT sequencing, which reveals kilobases of sequence in a single read, can provide more exact information than conventional sequencing methods (10). In the present V113 isolate, 2 beta-lactamase encoding genes (blaKPC and blaCTXM) were found on the plasmid. The blaKPC genes that encode KPCs are generally present on transferable plasmids and are flanked by transposable elements, thereby allowing for the gene to move from plasmid to the bacterial chromosome and back.
In conclusion, meropenem combined with ertapenem might be used for treatment of patients with ACLF and multi-drug resistant K. pneumoniae V113 isolate infection.