1. Background
Dilated cardiomyopathy (DCM) is among the most prevalent causes of heart failure and sudden cardiac death worldwide (1). The prevalence of DCM is four in 10,000 people, 30 - 50% of whom are afflicted with familial dilated cardiomyopathy (2). Despite immense medical advances in the last decades, such as the feasibility of heart transplantation and the availability of proper pharmaceutical agents, the prognosis of this disease has not enhanced significantly; therefore, early diagnosis and prevention of the progression remain the best option (3). Previously, environmental factors were attributed to the pathogenesis of this disease; in addition, recently, the genetic predisposition of the disease with the Mendelian mode of inheritance (mostly autosomal dominant) has been proven (4). Familial DCM is clinically and diagnostically the same as other forms of DCM; consequently, careful attention to family history is essential in the approach to DCM patients. The diagnosis of familial DCM is based on having one relative diagnosed with DCM or a history of sudden unexplained death in one of the first-degree relatives aged under 35 (5, 6). More than 50 genes have been discovered to be involved in the pathogenesis of familial DCM (7). Mutations in the nucleophilic A-type lamins, including the lamin A and C (LMNA) gene located on the long arm of chromosome 1, account for 5% to 8% of familial forms of DCM (8). According to the latest reports by American Heart Association (AHA), this gene is believed to have a definitive role in the DCM phenotype (9). LMNA protein coding gene has 12 exons in which a defect can cause DCM with or without conductive system abnormalities (10). It is the coding section for lamin-A and lamin-C proteins in the protein network of the inner nuclear membranes of the non-proliferative cells. Coding unfunctional lamin-A and lamin-C might impair nuclear membrane function, which would cause myocytes’ disintegration and destruction of the cardiac tissue (11). More than forty different mutations in the LMNA gene are associated with various conditions such as autosomal dominant Emery-Dreifuss muscular dystrophy, limb-girdle muscular dystrophy, familial DCM, and familial partial lipodystrophy (12, 13). Rs505058 polymorphism is an alteration in the Lamin gene’s third organic base of the 446th codon located on exon seven, by which a cytosine base substitutes thymine. This single nucleotide polymorphism (SNP) is one of the most common mutations seen in familial DCM (14, 15).
Is this SNP seen in patients with DCM in the Iranian population or not? Knowing these correlations might be helpful in future gene-based treatments and early diagnosis of this disease.
2. Objectives
We designed this study to evaluate the association of rs505058 T/C SNP in the LMNA protein-coding gene with familial dilated cardiomyopathy in Iranian patients.
3. Methods:
3.1. Study Design
The study followed a case-control protocol. Seventy DCM patients with at least one relative diagnosed with DCM or any unexplained sudden cardiac deaths in their first-degree relatives younger than 35 were enrolled in the study as the case group. Seventy healthy participants with normal left ventricular ejection fraction (LVEF) matched to the case group regarding their sex and age entered the study as the control group.
Written informed consent was obtained from all participants. The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of Jahrom University of Medical Sciences, Iran. All the individuals in the case and control groups underwent detailed history taking, physical examination, 12-lead electrocardiography, and transthoracic echocardiography. The considered echocardiographic criteria for diagnosis of dilated cardiomyopathy were as follows (14):
(1) Fractional shortening < 25% (reduction of > 2 standard deviations) and/or l LVEF < 45% (reduction of > 2 standard deviations);
(2) Left ventricular end-diastolic diameter more than 117% of the predicted value for age and total body surface area.
The study’s inclusion criteria were: Diagnosis of DCM in the patient and at least one of their first-degree family members by the mentioned criteria.
The study’s exclusion criteria were: Lack of proof for DCM, incompliance with the patients, unwillingness to enter the study, and the lack of family history for DCM.
3.2. Genetic Study
A 5 to 20 cc of the participants’ median cubital vein blood was collected in tubes containing EDTA to avoid clot formation. Then deoxyribonucleic acid (DNA) was extracted from the nucleated blood cells according to the instructions of the kit company (Sinagen Co., Iran). DNA was quantified spectrophotometrically prior to being used in the polymerase chain reaction (PCR). Next, extracted DNA was stored at -20°C before the PCR. The lamin gene sequence containing the rs505058 polymorphism was investigated using PCR amplification with specific forward GAGATCCACGCCTACCGCAAG and Reverse primer: AGCCAAAGAGTCCAGGAGCCA (Table 1).
| Sequences | Primer |
|---|---|
| 5’-GAGATCCACGCCTACCGCAAG-3’ | Forward |
| 3’-AGCCAAAGAGTCCAGGAGCCA-5’ | Reverse |
PCR reactions were performed in master mix tubes (Amplicon, Denmark). The reaction volume of 20 μL contained template DNA 2.5 μL, primers each 10 pmol, magnesium chloride 1.5 μM, dNTP Taq polymerase enzyme 250 μM each unit, and sterile water to 20 μL total reaction volume. The PCR reaction was done using a thermocycler (thermal cycler). The PCR program was as follows: 94°C for 5 min, with 35 cycles of 94°C for 40 seconds, 61°C for 30 seconds, and a final extension at 72°C for one minute and 72°C for 10 minutes, and ended. Then, tube samples were stored at -20°C. Gene runner software was used to determine the duplicated parts of primers, and gene sequences were confirmed by the blast program. The primers yielded a PCR product of 533 base pairs (bp) which were wholly digested with the restriction endonucleases Ear I for 24 h at 37°C to detect the rs505058 (T/C) polymorphism. Amplification products with the T allele (wild type) were digested into 219 and 314 bp DNA fragments with the restrictive enzyme Ear I (TT genotype and CT genotype). But the homozygote genotype CC did not undergo digestion with the enzyme. DNA fragments obtained after restrictive enzyme digestion and the DNA size marker were electrophoresed on a 3% gel agarose and stained with ethidium bromide.
3.3. Statistical Analysis
The data were registered into IBM SPSS for Windows, version 21.0 (SPSS Inc. Released 2007. Chicago, IL, USA) for statistical analysis. The quantitative variables were reported as mean, and standard deviation, and the qualitative variables were reported as frequency and percentages. A chi-square test was used to evaluate the association of rs505058 SNP with familial DCM. P-values less than 0.05 were counted as statistically significant.
4. Results
A total of seventy cases and seventy controls entered the study. Unfortunately, five samples of the case group were missed during the PCR and electrophoresis procedures which were withdrawn from statistical analysis.
The mean age of the participants was 28.87 ± 15.30 years for the case group and 29.18 ± 15.409 years for the control group. The age difference between the two groups was not statistically significant (P-value = 0.906).
In the control group, 57.4% (n = 40) of the participants were male, and 42.6% (n = 30) of them were female, while in the case group, 53.2% (n = 35) of the participants were male and 46.8% (n = 30) of the participants were female. The sex difference between the two groups was not statistically significant (chi-square test P-value = 0.700) (Table 2).
| Variables | Control Group (n = 70) | Case Group (n = 65) | P Value |
|---|---|---|---|
| Gender | 0.700 | ||
| Male | 40 (57.1) | 35 (53.8) | |
| Female | 30 (42.9) | 30 (46.2) | |
| Age (y) | 29.18 ± 15.40 | 28.87 ± 15.3 | 0.906 |
a Values are expressed as No. (%) or mean ± SD.
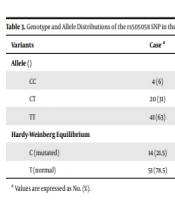
The Hardy-Weinberg equilibrium was used to survey the repartition of the rs505058 LMNA gene SNP genotypes, demonstrating a success rate of 100%. The wild-type TT genotype (P-value = 0.011) were significantly higher in the control group, while the mutated heterozygote genotype CT (P-value = 0.002) were significantly higher among the cases. Likewise, the mutated homozygote genotype CC was higher in the cases, but the relation was not statistically significant (Table 3). Considering the allele level comparison, the LMNA rs505058 C allele was accompanied by an enhancement in the incidence of dilated cardiomyopathy (OR = 2.92; 95% CI = 1.05 to 8.15, P-value = 0.034).
a Values are expressed as No. (%).
5. Discussion
The correlation between SNPs of the LMNA gene and DCM had yet to be previously studied in Iran; However, this study is the first study on the rs505058 polymorphism of the LMNA gene in Iranian population (16, 17). We found that the prevalence of rs505058 T/C SNP of the LMNA gene was higher in DCM patients than in healthy individuals in the Iranian population. C allele has shown a significant direct relationship with the prevalence of familial DCM in our studied population. In addition, the mutated heterozygote CT and homozygote CC genotypes prevalence were higher among the DCM patients compared to our controls. However, this relation was not statistically significant for the homozygote CC genotype, possibly due to the small sample size. Our findings were in line with previous works conducted on this issue worldwide. The higher rates of this mutation in patients indicate a direct relationship between this genetic alteration and the pathogenesis of DCM; however, the exact mechanism of this relationship is yet to be found.
Dilated cardiomyopathy is a disease of cardiac myocytes characterized by systolic and diastolic dysfunction. DCM can be presented as heart failure, thromboembolism, and sudden cardiac death (18, 19). Currently, mutations in more than 50 genes have been found to be associated with autosomal dominant DCM. LMNA gene encodes nuclear envelope proteins lamin A and lamin C, located on chromosome 1, and is one of the strong and definitive associating genes (20). Patients with symptomatic DCM with LMNA mutations seem to have a poor prognosis and carry a greater risk of complications such as dysrhythmias, sudden cardiac death, and heart failure than mutations in other loci (21).
Fatkin et al. were the first to describe the association between LMNA mutations with FDC accompanied by conductive system abnormalities such as sinus bradycardia, atrioventricular blocks, or atrial arrhythmias (10). Conductive system abnormalities might prompt the need for pacemakers and can become life-threatening in longstanding unattained LMNA-related DCM (22). Arbustini et al. introduced K97E, E111X, R190W, and E317K mutations of LMNA to be associated with FDC (13). They reported that around one-third of patients with DCM atrioventricular blocks carry mutations in their LMNA gene (13). Patients with symptomatic DCM with LMNA mutations have a poorer prognosis and take a greater risk of systolic and diastolic dysfunction, thromboembolic events, and sudden cardiac death than mutations in other loci (18). In patients with LMNA mutations, cardiac arrhythmias such as atrial fibrillation and ventricular tachycardia might be the first presenting symptoms long before the progression of DCM. It is yet to be evaluated if antiarrhythmic pharmacotherapy has a role in decreasing the pace or halting the process of the pathogenesis of DCM (12).
Gupta et al. reported that large deletions in the LMNA gene could cause insufficiency of lamin A/C proteins, thus disrupting the typical architecture of the nuclear envelope; however, they could not find any relations between SNPs of the LMNA gene and the severity of DCM (8).
Recently many studies have focused on the association between different SNPs of the LMNA gene and the pathogenesis of DCM (22-25). In rs505058 SNP, an alteration from thymine to cytosine in the 3rd base of codon 446 results in a nonsense variation to Aspartic acid at exon 7 of the LMNA gene (26, 27). Banerjee et al. reported the linkage between rs505058 SNP of the region encoding the central rod domain of lamin A/C proteins in an Indian population in 2015 (14). Sylvius et al. evaluated the relationship between two distinct LMNA mutations with cellular alterations (20). They found that the D192G mutation, in contrast to the R541S mutation, was associated with significant changes in the ultra-structure of the nuclear envelope. Furthermore, patients with D192G haploinsufficiency had a dimmer prognosis than patients with heterozygous R541S mutation. They also speculated that such mutations interfere with sumoylation-dependent cellular activities such as RNA transcription (20). Such underlying mechanisms might be involved in the pathogenesis of familial DCM associated with rs505058 SNP. However, these theories need further evaluation with more extensive studies.
Another proposed underlying mechanism for the pathogenesis of LMNA-related DCM comprises defective chromatin organization in the proliferating cells and signal transduction of non-proliferating cells (10, 20). Furthermore, LMNA mutations were found in more than 85% of patients with concurrent DCM and muscular dystrophy and in one-quarter of patients with isolated conductive system abnormalities, which prompts further evaluation of the genetic causes of conduction system disease (28).
5.1. Limitations
This study should be done in multi centers from different parts of the country, enrolling more participants. Moreover, in this study, long-term follow-up of the patients was not achieved, which can be counted as a limitation.
5.2. Conclusions
We conclude that the rs505058 T/C SNP in the LMNA gene is associated with the higher incidence of familial DCM among the south of Iran population; however, due to the limited number of participants in this study, further research is encouraged.