1. Background
Thyroid cancer is the fourth most common cancer worldwide. Its incidence has increased over the last decades. Based on the Surveillance, Epidemiology, and End Results program (SEER) data, the number of new cases of thyroid cancer were 52,070 and the percentage of all new cancer cases were 3% in 2019 (http://seer.cancer.gov/statfacts/html/thyro.html). This rate will increase to 52,890 new cases at the end of 2020 (1). Iran Cancer Data System Registry has reported 10,913 cases of thyroid cancer (7 - 90 years old) while the total incidence rate (per one year) was 2.20 per 100,000, during 2004 - 2010 (2). These findings show the increased rate of thyroid cancer in recent years. Thyroid carcinoma is classified into four groups; papillary thyroid carcinoma, follicular thyroid carcinoma, medullary thyroid carcinoma and anaplastic type. papillary thyroid carcinoma is the most common type of thyroid cancer accounting for about 80% of thyroid neoplasms (3, 4).
B-Raf proto-oncogene serine/threonine kinase (BRAF) mutations, especially V600E, are the most important genetic factors, being responsible for different aggressive phenotypes such as angiogenesis and tumor invasion (5). In 2016, the American Thyroid Association (ATA) guidelines suggested that BRAF and telomerase reverse transcriptase (TERT) promoter mutation are valuable molecular prognostic markers in PTC cases (6). In addition to genetic mutations, epigenetic modifications are also exemplified in explaining the initiation and progression of cancer. Nowadays, epigenetic changes in addition to genetic changes are responsible for the initiation and progression of cancer through activation of proto-oncogenes or inactivation of tumor suppressor genes (7). Promoter hypermethylation of tumor suppressor genes and/or global hypomethylation play an important role in thyroid carcinoma. Promoter hypermethylated tumor suppressor genes can be considered as an attractive candidate in the development of cancer biomarkers (8-10). Aberrant promoter methylation of CpG islands upstream of any gene may diminish transcription leading to decreased gene expression. On the other hand, DNA global hypomethylation could affect genomic stability (11, 12).
In 1998, Ueki et al. (13) identified novel nucleolar protein, termed NOLP (Nucleolar Localized Protein) which is now known as nucleolar protein 4 (NOL4). In spite of a number of different studies on the role of NOL4 as a tumor suppressor gene in different types of cancer like squamous cell carcinoma of head and cervical cancer, cervical cancer and Pancreatic ductal adenocarcinoma (7, 14, 15), there has been no study on the role of this gene in thyroid cancers to date.
2. Objectives
In the current study, we aimed to investigate whether expression and methylation of NOL4 are altered in PTC tissues in comparison with multi nodular goiter (MNG) tissues. Also we aimed at finding out whether NOL4 could be considered as a tumor suppressor gene in thyroid cancer.
3. Methods
3.1. Patient Recruitment and Sample Collection
Ninety candidate patients for thyroidectomy were recruited in this study. Written informed consent was obtained from each patient before sample collection. Thyroid tissues were resected during thyroidectomy and were snap-frozen in liquid nitrogen immediately after resection. Based on the pathology report, tissue samples of 41 PTC patients and 38 MNG patients were collected. B-RafV600E mutation was assessed using V600E‐B‐raf Q‐PCR kit (Cat no.: G001; HumDiagnostics, Iran). This study has been approved by the Ethics Committee of the Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Science (ethics code: IRI.SBMU.ENDOCRINE.RES.1397.067).
3.2. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted using RNX-Plus solution for total RNA isolation according to the manufacturer’s protocol (Cinnagen, Iran, Cat. no.: RN7713C). RNA purity and concentration were detected by NanoDrop-1000 (Thermo Fisher Scientific). RNA was converted to cDNA using the RevertAid Reverse Transcriptase (ThermoFisher Scientific, Cat no.: MAN0012757) according to the previously mentioned protocols (16). Quantitative RT-PCR analysis of NOL4 expression was conducted in triplicate using Real Q Plus 2X Master Mix Green (Ampliqon, Cat. no.: A325406) on the ABI Step One Plus System (Life Technologies, USA). GAPDH level was studied as an internal control. Primers were designed by Primer3 online tool. Primer sequences for NOL4 and GAPDH are shown in Table 1. Relative mRNA expressions were normalized using average of ΔThreshold Cycle (ΔCT= CT target gene-CT GAPDH). Expression of this gene in each PTC sample was compared with the average expression in MNG samples one by one. Fold change of expression was estimated using 2-ΔΔCT while ΔΔCT stands for ΔCT of MNG cases subsided by ΔCT of PTC cases. Expression data was divided into low expression (fold change ≤ 0.5), unchanged expression (fold change ranged from 0.5 to 2), and over expression (fold change ≥ 2) based on a published article by Rismani et al. (17).
| Gene Name | Forward Primer Sequences | Reverse Primer Sequences | Ta | CpG Sites (n) | Bases (n) |
|---|---|---|---|---|---|
| Primer Sequences for MS-HRM for Studying Methylation | |||||
| NOL4 | |||||
| Locus a | GTTGTTTTAAGGTTAGATTTAGAGG | CCAATCTCTCTAATTACCCAATAA | 57 | 11 | 250 |
| Locus b | GTAGGATTTATTGATTATTTTGTAAGTG | GCTCCTCTTTATTCTACTGTCACC | 51 | 20 | 223 |
| Locus c | GGTGAGAGTAGAATAAAGAGGAG | ATCATTTTCCCGTTACTTAATCAC | 57 | 8 | 240 |
| Locus d | TAGAAATTAGAGAAGTTTTGTTTGG | TTCTTACCTATCATATTTTTAAACCTATC | 55 | 8 | 320 |
| Locus e | AAAGTAGGATTTAGAAGGAAGGAGAG | CCAACACGATTCTAACCCAAAAAAAC | 59 | 21 | 338 |
| Locus f | TAGTTTTTTTGGGTTAGAATGGTGTTG | ACCCTAAACTCATAAAAAGTACAACCC | 59 | 13 | 278 |
| Primer sequences for RT-PCR | |||||
| NOL4 | GGCGAGAGAAATTGGAAGCA | CTCCGAATCGTCATGGTCCT | 59 | 151 | |
| GAPDH | TCGTGGAAGGACTCATGACC | AGGCAGGGATGATGTTCTGG | 60 | 151 | |
Abbreviation: CpG, cytosine–phosphate diester-guanine; MS-HRM, methylation sensitive high resolution melting; RT-PCR, real time polymerase chain reaction; Ta, appropriate annealing temperature.
3.3. DNA Extraction and Bisulfite Treatment of DNA
DNA of the samples was extracted using Tissue Genomic DNA Extraction Mini Kit (Favorgen, Cat no.: FATGK001). The quality and quantity of DNA samples were assessed using gel electrophoresis and NanoDrop-1000 (ThermoFisher Scientific, USA). DNA was bisulfite converted with the EZ DNA Methylation-Gold Kit (Zymo Research, USA, Cat no.: D5006) according to the manufacturer’s protocol.
3.4. Methylation-Specific High Resolution Methylation (MS-HRM) Assay
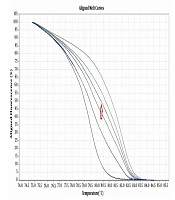
Six pairs of specific primers were designed for studying the methylation level of NOL4 promoter regions which were named from NOL4 (a) to NOL4 (f). Primers were designed according to bisulfite modified sequence and they should cover at least 8 CpG islands within the amplified region through Gene Runner version 6.5.5 and AlleleID 6 softwares. Meth Primer site (www.urogene.org/methprimer) was used to find CpG islands in NOL4 promoter region (Table 1). MS-HRM experiments were performed in ABI Step One Plus System (Life Technologies, USA). Reaction mixtures consisted of 5 µL of Real Q Plus 2X Master Mix Green (Ampliqon, Cat. no.: A325406), 10 pmol of each primer and 1 µL (equal to 10 ng) of bisulfite modified DNA template in the final volume of 10 µL. Standard samples including methylated and non-methylated human DNA (Hum Diagnostics, Iran) were prepared as described in Refs (18). Five standard curve includes; 0%, 25%, 50%, 75%, and 100% methylation levels were used for detecting the methylation level of each sample. The highest curve was 100% methylation level, and the lowest one was 0% methylation level. The arrow was showed one sample methylation level, which was located between 50% and 75% methylation levels (Figure 1).
3.5. Statistical Analysis
Normal distribution of data was evaluated by employing the Kolmogorov-Smirnov test. The methylation and expression differences with and without normal distribution, were determined by Independent samples t-test and Mann Whitney U test, respectively. Correlation analyses were done using Pearson, Spearman and chi-Square tests. Receiver operating characteristic curves was utilized to determine the diagnostic value of tests. Furthermore, area under the curve (AUC), threshold, specificity and sensitivity were calculated. The accuracy of AUC classification for a diagnostic test was as follows; AUC between 0.9 - 1.0, 0.8 - 0.9, 0.7 - 0.8, and 0.6 - 0.7 are classified as excellent, good, fair, and poor, respectively (19). All P values less than 0.05 were considered statistically significant. All statistical analyses were performed using SPSS Version 22.0 (IBM Corporation, Armonk, NY, USA) and GraphPad Prism version 8 (GraphPad Software, Inc., La Jolla, CA, USA).
4. Results
4.1. Demographic and Clinic-Pathologic Data of Patients
Demographic data for each group was considered as; PTC group (3 males, 38 females, 41.18 ± 11.41 years old), and MNG group (8 males, 28 females, 45.50 ± 13.27 years old). There is no significant difference in age and sex distribution between the two groups. Participants were mostly female (85.7%). The age ranges of PTC and MNG cases were 25 - 64 and 21 - 75 years, respectively. While twenty-six samples were in stage I, only six, four and one PTC samples were in stage II, III, and IV, respectively. Tumor size varied from 0.4 to 5.7 cm. Twenty-one PTC tumors were unifocal while 14 tumors were multifocal. Pathology reports of four cases were not available. Hashimoto’s thyroiditis was detected in 13 (35.1%) of the PTC patients. While 62.2 % of patients had invasion, BRAFV600E mutation was detected in 63.4% PTC patients.
4.2. NOL4 Was Downregulated in PTC Samples Which Was Affected by Methylation of Its Proximal Promoter Region
The methylation status of six regions which covers 81 CpG islands upstream of NOL4 gene, was analyzed using HRM assays. This analysis revealed that 38 out of 41 (92.68%) of PTCs and 33 out of 36 (91.6%) of MNGs did not show methylation in NOL4 (a) region. In this analysis non-methylation and methylation were defined as a methylation level less than 12.5 and methylation level more or equal to 12.5 respectively. The median methylation percentage of NOL4 (b) was approximately equal in PTC and MNG cases, whereas 12.19% PTC cases in comparison with 8.3% of MNG cases were methylated in this region. The NOL4 (c) gene was methylated in (27/41) PTCs and (27/36) MNGs. The methylation of NOL4 (d) occurred in 56.1% PTC cases in comparison to 22.2% of MNG cases. The methylated NOL4 (e) region was observed in 53.6% and 38.9% of PTC and MNG cases respectively. As it represented in Table 2, the median percentage of methylation in NOL4 (a/d) were 5 ± 0.9/5 ± 2.11 and 8 ± 1.17/10 ± 2.37in PTC and MNG tissues respectively which is slightly lower in PTC tissues. The median percentage of methylation in NOL4 (c/e) were 25 ± 4.38/12.5 ± 2.06 and 20 ± 3.57/10 ± 3.07 in PTC and MNG cases respectively. There is no difference in the methylation percentage of NOL4 (b) in both groups. Although, different methylation rates between PTC and MNG were observed in the above mentioned regions, they were not statistically significant. Methylation of NOL4 (f) (the nearest region to the start codon) was significantly higher in PTC cases compared to MNG cases (P value = 0.02). Maximum percentage of methylation in this region was 80% in PTC cases while this rate was 12.5% in MNG cases. While 10 PTC cases (24%) were methylated, only two MNG cases (5.5%) showed methylation in this region (Table 2).
| Gene Methylation | P Value | ||||||
|---|---|---|---|---|---|---|---|
| PTC Cases | MNG Cases | ||||||
| Median ± SE | Min-Max | Frequency | Median ± SE | Min-Max | Frequency | ||
| NOL4 (a) | 5 ± 0.90 | 0 - 25 | NM: 38; M: 3 | 8 ± 1.17 | 0 - 30 | NM:33; M:3 | 0.72 |
| NOL4 (b) | 5 ± 0.94 | 0 - 25 | NM: 36; M: 5 | 5 ± 1.20 | 0 - 35 | NM:33; M:3 | 0.59 |
| NOL4 (c) | 25 ± 4.38 | 0 - 100 | NM: 13; M: 27 | 20 ± 3.57 | 0 - 90 | NM: 9; M: 27 | 0.74 |
| NOL4 (d) | 5 ± 2.11 | 0 - 50 | NM: 26; M: 23 | 10 ± 2.37 | 0 - 50 | NM: 23; M: 8 | 0.89 |
| NOL4 (e) | 12.5 ± 2.06 | 0 - 80 | NM: 18; M: 22 | 10 ± 3.07 | 0 - 80 | NM: 22; M: 14 | 0.24 |
| NOL4 (f) | 6 ± 2.25 | 0 - 80 | NM: 31; M: 10 | 5 ± 0.60 | 0 - 12.5 | NM: 34; M: 2 | 0.02a |
Abbreviation: M, promoter methylation equal or more than 12.5; MNG, multinodular goiter; NM, promoter methylation less than 12.5; PTC, papillary thyroid carcinoma.
aP value less than 0.05 was considered significant.
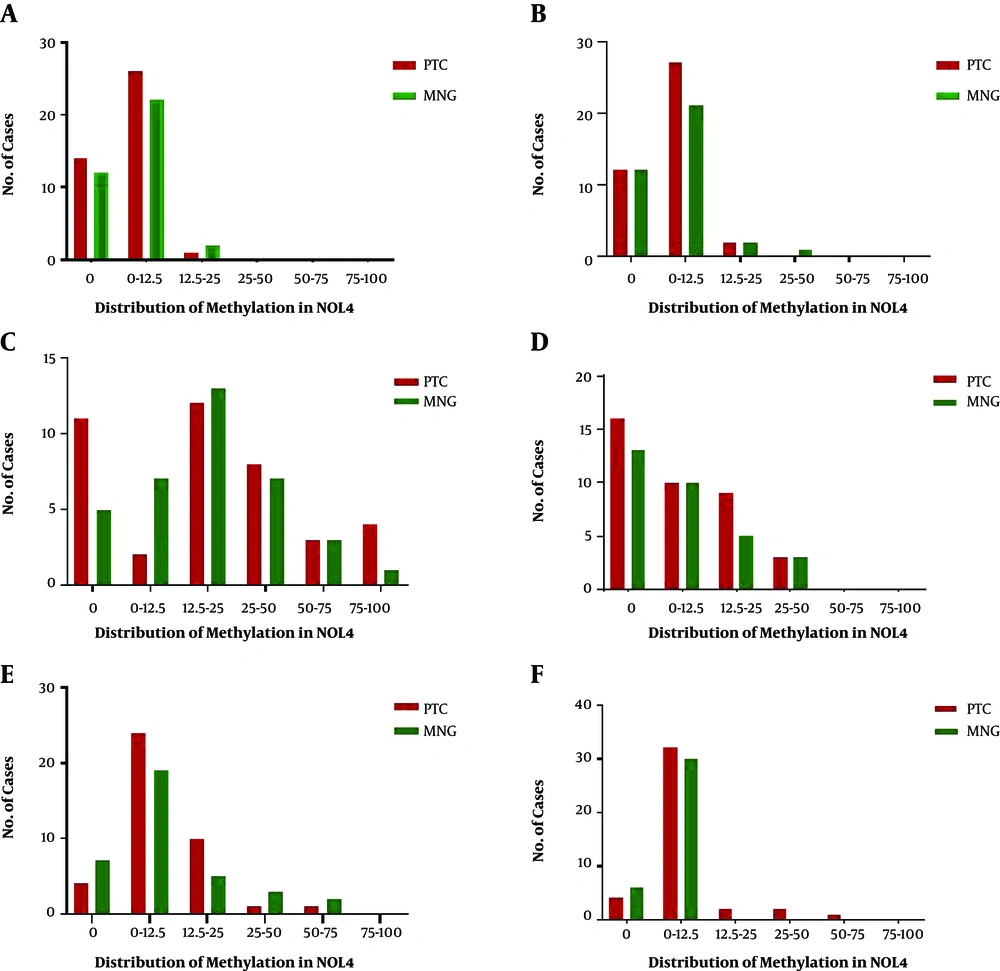
Methylation distribution is categorized in 6 classes (0, 0 - 12.5, 12.5 - 25, 25 - 50, 50 - 75, and 75 - 100) which are shown in Figure 2. The highest methylation level of NOL4 (a) ranged from 12.5% to 25%, which appeared in one PTC and two MNG cases. Methylation level of 25% - 50% in NOL4 (b) was detected in one MNG case. NOL4 (c) was methylated from 75% to 100% in four PTC cases, meanwhile one MNG case was methylated in this range. The highest methylation percentage (25 - 50) of NOL4 (d) was observed equally in both groups. Methylation levels of 50% and 75% in NOL4 (e) were observed mostly in MNG cases. Most of the MNG cases showed a methylation level of 12.5% in NOL4 (d) however, methylation levels of 25%, 50%, and 75% were solely detected in PTC cases. All methylation levels in different regions of NOL4 are significantly correlated with each other except NOL4 (d) which is solely correlated with methylation level of NOL4 (f).
Distribution of NOL4 promoter methylation in different regions; The methylation status are categorized into six classes; 0, 0% methylation; 0 - 12.5, methylation percentage < 12.5%. 12.5 - 25, methylation percentage from ≥ 12.5% to < 25%; 25 - 50: methylation percentage from ≥ 25% to < 50%; 50 - 75, methylation percentage from ≥ 50% to < 75%; 75 - 100, methylation percentage from ≥ 75% to 100%; PTC, papillary thyroid carcinoma MNG: multinodular goiter.
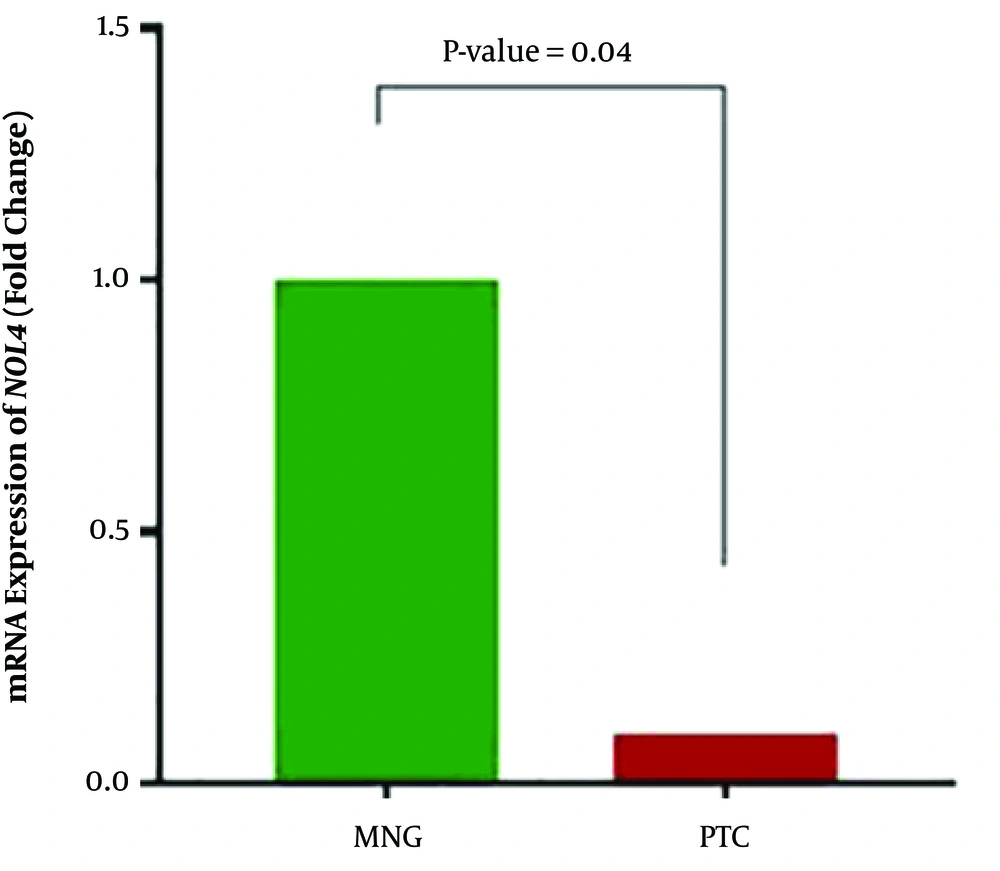
NOL4 expression was significantly downregulated in PTC cases compared with MNG cases (P value = 0.04). Downregulation of NOL4 was observed in 26 (66.7%) of the PTC cases while overexpression and unchanged expression were seen in 10 (25.6) and 3 (7.7%) of these patients, respectively. Mean relative fold change of expression was 0.1 in tumor samples (Figure 3). Spearman analyses revealed that in PTC samples, NOL4 expression was only inversely correlated with methylation in NOL4 (f) (correlation coefficient: -0.403, r2: 0.04, P value = 0.01). It means that downregulation of NOL4 is correlated with methylation of the most proximal region in the promoter of the NOL4 gene, suggesting the role of aberrant promoter methylation on NOL4 expression in PTC cases.
4.3. Methylation Status of NOL4 (f) Was Correlated with the age of Patients and Lymph Node Metastasis
Methylation status of NOL4 (f) directly correlated with age of diagnosis (age of the patient at the time of diagnosis) (correlation coefficient: 0.359, P value = 0.02). Gender, tumor size, extra thyroid invasion, vascular invasion, and capsular invasions, lymph node metastasis, clinical stage, and BRAF mutation did not have any significant correlation with NOL4 (f) promoter methylation. On the other hand, the correlation between the NOL4 (f) methylation status and clinic-pathological characteristics was also analyzed using chi-square analysis and univariate logistic regression analysis. Lymph node metastasis (OR = 0.122, 95% CI = 0.013 - 1.14; P = 0.05) was associated with the methylation of NOL4 (f). On the other hand, no correlation was found between NOL4 (f) methylation status and age of diagnosis, gender, tumor size, extra thyroid invasion, capsular invasions, clinical stage, previous history of Hashimoto’s thyroiditis, and BRAFV600E mutation. Methylation level of other regions in NOL4 promoter showed no remarkable correlation with clinic-pathological features of disease.
4.4. NOL4 is Downregulated in Advanced Stages of Disease
NOL4 mRNA expression was significantly correlated with the stage of disease (correlation coefficient = 0.35, P value = 0.03) though age of diagnosis, gender, BRAF mutation, and other clinic-pathological features were not significantly associated with NOL4 mRNA level. On the other hand, the chi-square analyses revealed no relationship between NOL4 expression status and age, sex, tumor size, invasions, lymph node metastasis, clinical stage, previous history of Hashimoto’s thyroiditis, and BRAF mutation.
4.5. NOL4 mRNA Level Could be A Poor Diagnostic Markers for PTC
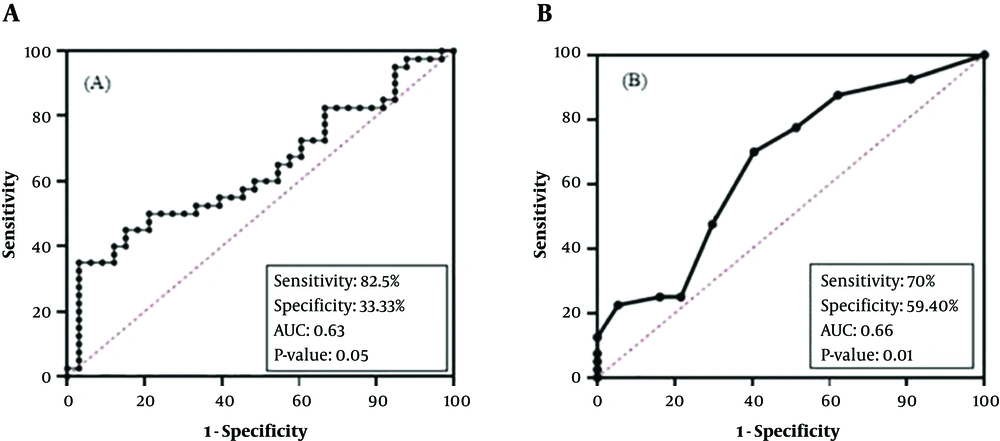
ROC curve analysis was used for determining the diagnostic value of biomarkers which were significantly different between PTC and MNG cases (Figure 4). NOL4 mRNA level showed significant differences at a cut-off of ΔCT > 3.38 [sensitivity: 82.5%, specificity: 33.3%, and an AUC: 0.63] in PTC cases. (95% CI = 0.5 - 0.7, P value = 0.05).
4.6. Methylation of NOL4 (f) Region Can be Considered as a Poor Diagnostic Marker for PTC
The promoter methylation of NOL4 (f) had a diagnostic value in cut of point of methylation level more than 5.50% [sensitivity: 70%, specificity: 59.40 % (and AUC = 0.66 (95% CI = 0.5 - 0.8, P value = 0.01)]. In regard to AUC, this marker showed poor diagnostic value.
5. Discussion
In the present study, in PTC and MNG samples the methylation status of promoter regions of NOL4 were investigated by MS-HRM. Methylation of the most proximal region in NOL4 promoter was observed in PTC patients. This observation was in accordance with the expression level of this gene in these samples suggesting a possible tumor suppressor role of this gene. Since discovery of NOL4 gene which is located on 18q12.1 chromosome (13), there have been limited studies focusing on the role of NOL4 in tumorigenesis. Wang et al. (14) identified aberrant methylation of NOL4 in cervical cancer. Meanwhile, Demokan et al. (7) reported that aberrant methylation of CpG islands upstream of this gene was associated with head and neck squamous cell carcinoma (HNSCC). Furthermore, one study recently revealed that the NOL4 gene plays a role in glioblastoma multiform (GBM) (15). NOL4 intronic mutation has been detected in Indian families with Van der Woude syndrome (20). Based on the NOL4 expression in normal thyroid tissues (21) and lack of sufficient evidence on the tumorigenesis effect of NOL4 in thyroid cancer, we decided to investigate this study. In this study, it was shown that in PTC patients, NOL4 downregulation is associated with aberrant promoter methylation of this gene. We also found correlation of NOL4 gene methylation and expression with lymph node metastasis and advanced tumor stages, providing strong evidence to support the role of NOL4 as a tumor suppressor gene in papillary thyroid carcinoma and it is recommended it be validated in future studies. To date, NOL4 molecular functions remain unclear but it is known that NOL4 is expressed during facial (lip and palate) development and brain and testis in humans and mice (13, 20). NOL4 hypermethyation in different cancers especially cancer located in head and neck support the hypotheses that it can act as a tumor suppressor gene which should be investigated in further experiments.
In this study, NOL4 methylation in addition to downregulation of it are introduced as novel biomarkers of PTC though the sensitivity of NOL4 methylation was lower to be ideal markers. In order to confirm these findings, a larger sample size from patients affected with PTC in different stages should be recruited. In addition, studying the methylation and expression of these genes should be assessed in other types of thyroid cancer or other types of cancer too. On the other hand, study on the effects of NOL4 on histone modification could help to better understanding of its epigenetic function.
In conclusion, this was the first study to investigate the role of the NOL4 gene in PTC patients. NOL4 was significantly methylated and downregulated in PTC patients. While methylation of NOL4 was associated with lymph node metastasis, downregulation of this gene was linked with advanced stages of PTC. So, the present study provided strong evidence to support the hypothesis that NOL4 could be a putative tumor suppressor in papillary thyroid carcinoma.