1. Background
Non-communicable diseases and notably cardiovascular diseases (CVD) are the primary reason for mortality and the main health burden worldwide (1). Serum lipids and lipoproteins in the pediatric age group are shown to be predictive of future symptomatic CVDs (2). Studies show that lipoproteins, as well as other cardiovascular risk factors, track from childhood to adulthood (3). The assessment of the early onset of CVD risk factors in childhood is important to determine optimal preventive measures, since determining the early development of adult CVDs may help reduce the mortality rate (4, 5). Among lipids, high-density lipoprotein cholesterol (HDL-C) is a protective element against CVD (6). A study presented that having HDL-C 6 to 7 mg/dL higher than average leads to 20% to 27% decrease in the risk of CVD (7).
Low HDL-C levels are quite prevalent in the Middle-Eastern countries (8). Population-based studies in Iran presented a prevalent low HDL-C among children and adolescents (4, 9, 10). Furthermore, its fifth percentile of Iranian pediatric population is lower than that of European and American population (11, 12). Moreover, it is well documented that HDL-C is affected by both genetic contexts and environmental factors, such as demographics, diet, smoking, and weight disorders (13, 14). Although hormonal, environmental, and social factors could specify HDL-C levels, the genetic component accounts up to 76% of the variation in HDL-C levels (15). Single nucleotide polymorphism (SNP) is the most prevalent genetic discrepancy (16). In the evaluation of genotype data, the effect size of associations between one SNPs and a response is usually small. Therefore, it is assumed that not only single SNPs but also interactions of several SNPs is particularly important (17). Most genetic studies on lipid profiles focused on individual SNPs while SNP-SNP interactions are suggested to have a great effect on the structure of complex diseases. As an example, Kelishadi et al. investigated the genetic association with low concentrations of HDL-C in a pediatric population, (18). In this study, single SNPs were taken into accounts and the polymorphism of ApoE gene was not included.
To the best of our knowledge, no analysis has been performed to evaluate an association between the interaction of SNPs and low HDL-C in Middle Eastern adolescents. Therefore, the aim of the present investigation was to determine the influence of some polymorphisms (and their interactions) on HDL-C levels performed for the first time on children and adolescents.
2. Methods
2.1. Study Populations
The subjects in this study were randomly determined between individuals in the CASPIAN-III study, Childhood and Adolescence Surveillance and Prevention of Adult Non-communicable disease study. This survey was performed to determine risky behaviors in Iranian school students (2009 - 2010).
The original study included 5528 10 - 18 year-old students selected from Iranian urban and rural regions (19). A total of 734 frozen blood samples were selected.
Low HDL-C levels were determined as concentrations of < 40 mg/dL. Elevated lipid was considered as total cholesterol higher or equal to 200 mg/dL, low-density lipoprotein cholesterol (LDL-C) higher or equal to 130 mg/dL, and triglycerides higher or equal to 150 mg/dL. High waist circumference was determined as WC higher or equal to 90th percentile of the population studied. Elevated fasting blood sugar was defined as FBS higher or equal to 100 mg/dL. Blood pressure higher or equal to 130/85 mmHg was considered as elevated BP (20).
Normal weight Underweight, overweight, and obesity were determined as a BMI Z-score according to world health organization (WHO) definition (21).
Polymorphisms of LPL rs1801177, LPL rs320, LPL rs328, ABCAI rs2066718, ABCAI rs2230808, CETP rs708272, CETP rs5880, APOC3 rs5128, APOA1 rs2893157, APOA5 rs662799, and ApoE genes related to HDL-C disorder were analyzed (22). Each SNP was represented by the values 0, 1, and 2 based on the pairs of nucleotide. We recoded this variable into 2 binary variables corresponding to a dominant gene (if SNP has at least one variant allele) and a recessive gene (if SNP has 2 variant alleles). Using this method, we generated 2p binary predictors out of p SNPs (23).
2.2. Genetic Studies
2.2.1. DNA Extraction
DNA was extracted from peripheral blood by using the QIAamp DNA Blood Mini kit (Qiagen, Germany). Corbett rotor-gene 6000 instruments (Corbett Research Pty Ltd, Sydney Australia) were used for Real-time PCR and high-resolution melt analysis. Primers were produced by Beacon Designer 7.91 to flank the genomic regions (PREMIER Biosoft International, USA, and TIB MOLBIOL (Germany) were used for synthesizing.
The Standard conditions of amplicons production using type-it HRMkit (Qiagen, Germany) were used (24). The sequence-proven major and minor allele homozygote and heterozygote controls were then included. The HRM analysis was performed, and the samples were clustered (24).
2.3. Statistical Analysis
The continuous variables were shown as mean and standard deviation. However, on the other hand, frequencies and percentages were used for categorical variables. The t-test and χ2 were used to compare the continuous and categorical variables in boys and girls, when appropriate. The association between polymorphisms and other covariates interactions and HDL-C levels was analyzed using logic regression.
Few approaches were proposed for the direct detection of statistical interactions between SNPs in order to enhance the statistical power of the studies. Logic regression was recently proposed as a generalized regression method to identify interactions between binary predictors associated with a response variable. It can detect complicated interactions between predictors that could play important roles in genetics or discover prognostic factors in medical data. This technique was efficiently applied on SNP data (23).
The form of the logic regression model is as below:

where h(.) is an appropriate link function for the response and the predictors, e.g. linear (h (E [Y|X]) = E [Y]), and logistic regression (h (E [Y|X]) = log(E [Y]/(1 - E [Y]))). X is the covariate matrix, with βi as parameters and the Li as Boolean combinations of the binary predictors such as x2 and x5c or x7, with the operator ‘c’ as complement. Furthermore, the Zj are extra confounders. For each model, a score function was defined reflecting the “quality” of the model under consideration. We endeavored to find Boolean combinations minimizing the scoring function related to the model. Simulated annealing search algorithm was used to search for the best Boolean combinations and estimation of βj (23).
The association between combination of predictors and low HDL-C level was assessed by the randomization test. The best model was first fitted to the data, the response was permuted in random, and the best model was re-fitted. This procedure was repeated several times. If the entire scores of the permuted data were considerably worse than that of the original data, the information existed in the predictors; otherwise, there was no connection between the predictors and the response (23).
To avoid over-fitting in logic regression models, the cross-validation test was applied to determine the optimal model size. The data were split into k equal parts. Then, k times, one partition was left as a test set, and for each possible model size, the best model was selected using k -1 out of k parts, after which the score function on the remaining test set part was computed. For each level of model size, the k score function was added and the model size with the smallest overall score function was selected (23).
Logic regression with logit link function was used to recognize the relationship between the input predators and the outcome. We applied R software (version 3.3.0) for analyzing the data.
3. Results
The average age of the adolescents was 14.66 (2.6) years with the male predominance of 51.6% (sex ratio = 1.068). Table 1 represents the summary of demographic and clinical characteristics of the study subjects by gender.
| Variable | Total (N = 734) | Boys (N = 379) | Girls (N = 355) | P Value |
|---|---|---|---|---|
| Age, ya | 14.66 ± 2.61 | 14.56 ± 2.62 | 14.76 ± 2.59 | 0.30 |
| BMI, kg/m2a | 19.14 ± 4.08 | 19.43 ± 4.20 | 18.82 ± 3.93 | 0.04c |
| HDL-C, mg/dLa | 49.50 ± 21.93 | 48.88 ± 21.23 | 50.16 ± 22.67 | 0.43 |
| High FBSb | 90 (12.26) | 35 (9.23) | 55 (15.49) | 0.01c |
| High WCb | 100 (13.62) | 49 (12.93) | 51 (14.37) | 0.57 |
| High TCb | 43 (5.86) | 21 (5.54) | 22 (6.2) | 0.71 |
| High LDLb | 58 (7.90) | 30 (7.92) | 28 (7.89) | 0.99 |
| High TGb | 64 (8.72) | 28 (7.39) | 36 (10.14) | 0.19 |
| Elevated BPb | 23 (3.13) | 10 (2.64) | 13 (3.66) | 0.43 |
Abbreviations: BMI, Body Mass Index; BP, Blood Pressure; FBS, Fasting Blood Sugar; HDL-C, High Density Lipoprotein Cholesterol; LDL, Low Density Lipoprotein Cholesterol; TC, Total Cholesterol; TG, Triglycerides; WC, Waist Circumference.
aValues are expressed as mean ± SD.
bValues are expressed as n (%).
cSignificant values.
Genotype and SNP allele frequencies were used to assess genetic association with HDL-C levels (Table 2). None of the SNP distributions represented the deviation from Hardy-Weinberg equilibrium.
| Polymorphism | Genotype and Allele | ||
|---|---|---|---|
| LPL rs1801177 genotypes | AA | AG | GG |
| 691 (94.1) | 43 (5.9) | 0 | |
| LPL rs320 genotypes | GG | GT | TT |
| 259 (35.3) | 336 (45.8) | 139 (18.9) | |
| LPL rs328 genotypes | CC | CG | GG |
| 580 (79) | 139 (18.9) | 15 (2) | |
| ABCAI rs2066718 genotypes | GG | GA | AA |
| 704 (95.9) | 30 (4.1) | 0 | |
| ABCAI rs2230808 genotypes | AA | AG | GG |
| 432 (58.9) | 248 (33.8) | 54 (7.4) | |
| CETP rs708272 genotypes | CC | CT | TT |
| 268 (36.5) | 371 (50.5) | 95 (12.9) | |
| CETP rs5880 genotypes | CC | CG | GG |
| 638 (86.9) | 92 (12.5) | 4 (0.5) | |
| APOC3 rs5128 genotypes | CC | CG | GG |
| 612 (83.4) | 118 (16.1) | 4 (0.5) | |
| APOA1 rs2893157 genotypes | GG | GA | AA |
| 529 (71.7) | 193 (26.3) | 15 (2) | |
| APOA5 rs662799 genotypes | CC | CT | TT |
| 722 (98.4) | 5 (0.7) | 7 (1) | |
| ApoE alleles | e2 | e3 | e4 |
| 31 (4.2) | 654 (89.1) | 49 (6.7) | |
aABCA1, gene encoding ATP binding cassette transporter A1; APOA1, gene encoding apolipoprotein (apo) A-I; APOA5, gene encoding apo A-V; APOC3 gene encoding apo C-III; CETP, gene encoding cholesteryl ester transfer protein; LPL, gene encoding lipoprotein lipase.
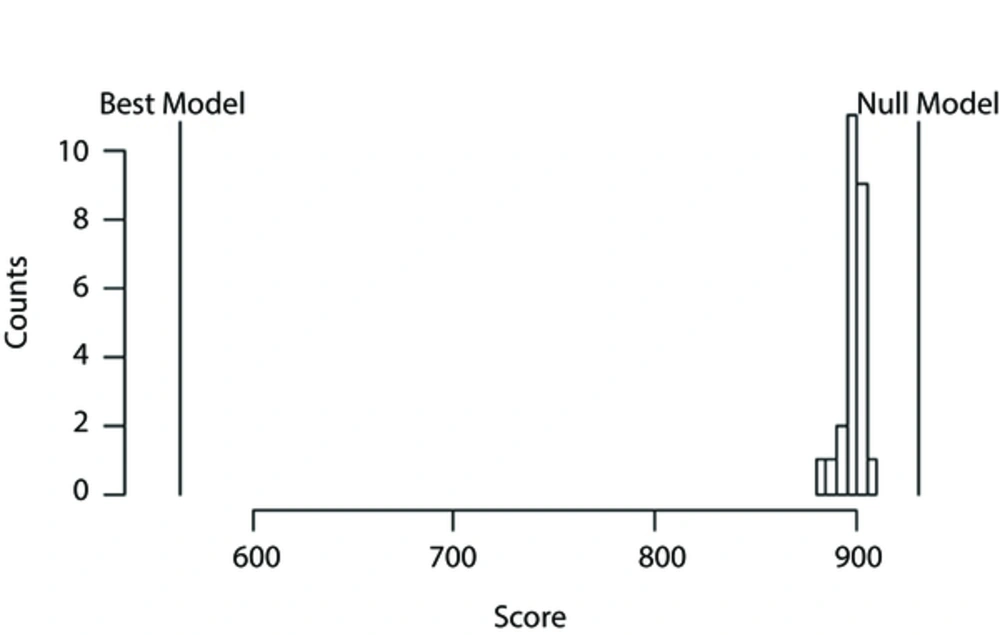
Figure 1 shows the results from the null model randomization test. The score of the NULL, the best scoring, and that of the randomization models were compared. Accordingly, we can conclude that there is an association between the predictors and the low HDL-C.
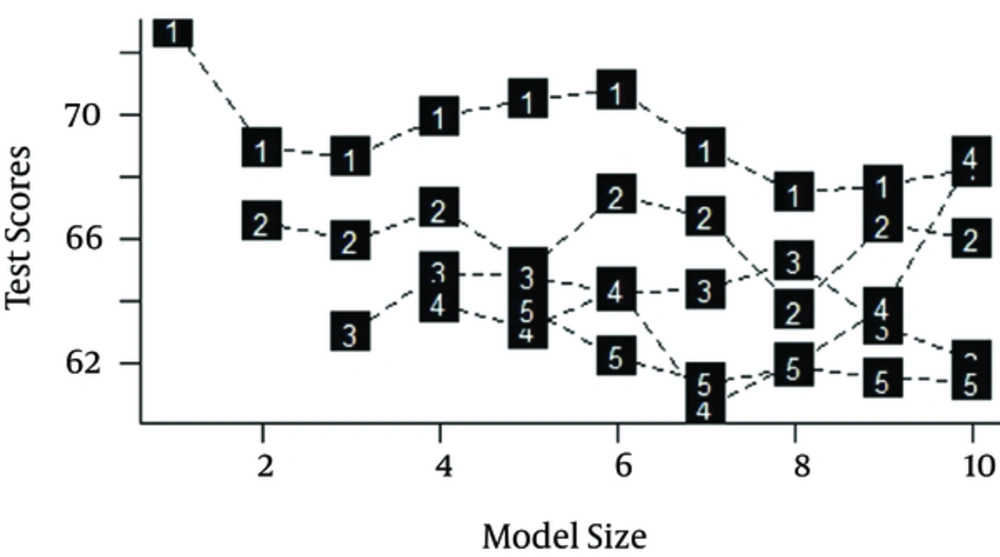
Figure 2 displays the result of the cross-validation test. Accordingly, the model with 4 trees and 7 leaves was optimal in terms of the test set deviance and the model size.
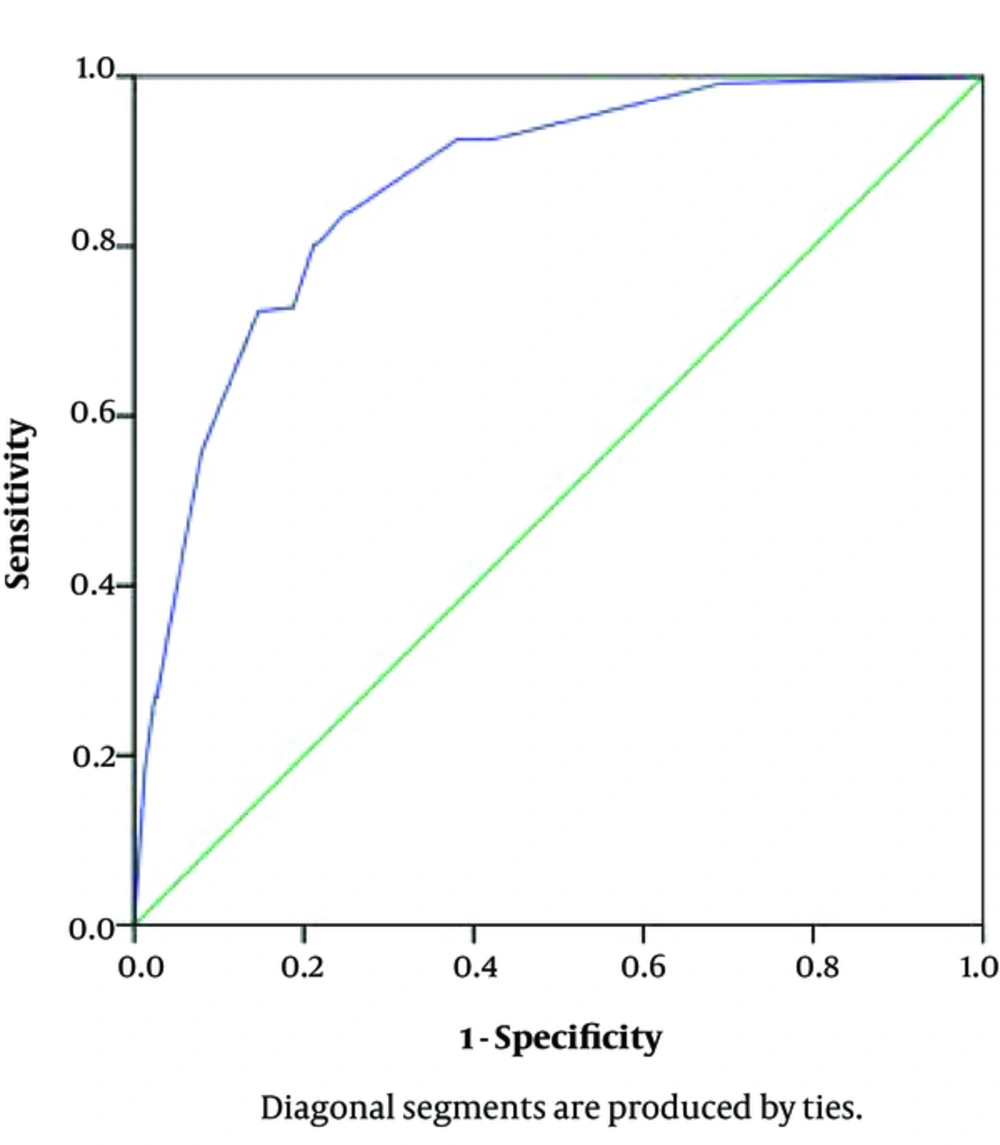
Figure 3 displays the ROC curve of fitted model. The Area under the ROC curve was 0.87 (95% CI 0.81, 0.90) indicating the high performance of the proposed method.
Table 3 presents the optimal combination rules for low HDL-C based on the logic regression. The model contains 4 logic combinations significantly affecting the low level of the HDL-C:
| Boolean Combination | Coefficient | Standard Error | Odds Ratio | 95% Confidence Interval | P Value |
|---|---|---|---|---|---|
| L1: ((rs2230808 = GG) or (rs5880 ≠ CC)) | 1.295 | 0.251 | 3.65 | (2.23, 5.98) | < 0.0001 |
| L2: ((rs320 = GG) and (rs1801177 ≠ AA)) | 0.749 | 0.211 | 2.12 | (1.4, 3.2) | < 0.0001 |
| L3: ((rs320 ≠ TT) or (rs708272 ≠ TT)) | 0.436 | 0.17 | 1.55 | (1.104, 2.165) | 0.011 |
| L4: (rs708272 ≠ CC) | -2.64 | 0.18 | 0.071 | (0.05,0.1) | < 0.0001 |
| AIC | 572.64 | ||||
| AUC | 0.87 |
Abbreviations: AIC, Akaike Information Criterion; AUC, Area Under the Curve.
The first combination (L1) contains “((rs2230808 = GG) or (rs5880 ≠ CC))”. This combination indicates that subjects with the GG genotype of ABCA1 rs2230808 or not CC genotype of CETP rs5880 have an odds ratio of 3.65 (95% CI 2.23, 5.98) with low levels of the HDL-C, comparing to the other cases.
The second combination (L2) is explained by the interaction of LPL rs320 and LPL rs1801177 polymorphisms. The estimated odds ratio for this combination was 2.12 (95% CI 1.4, 3.2), inferring that G allele of LPL rs1801177 and GG genotype of LPL rs320 gene is associated with low HDL-C levels.
The third combination (L3) contains “((rs320 ≠ TT) or (rs708272 ≠ TT))”. This form suggests that not TT genotype of LPL rs320 or not TT genotype of CETP rs708272 is associated with higher odds of low HDL-C levels. The odds ratio associated with L3 was 1.55 (95% CI 1.10, 2.17), indicating that, as a group, the subjects who complies with L3, are estimated to have higher odds of low HDL-C levels compared to the other subjects.
The fourth combination (L4) was entirely solely by CETP rs708272 polymorphism. The odds ratio related to this combination was 0.071 (9% CI 0.05, 0.1), inferring that T allele of rs708272 polymorphism is related with high HDL-C levels. The deviance as the model score function was 562.64.
4. Discussion
In this cross-sectional multi-center study, we investigated the effect of 11 genetic variants on the low HDL-C levels phenotypes and identified SNPs that were simultaneously associated with the HDL-C levels of adolescents. Although many studies examined the relationship of single SNP and low HDL-C levels, separately, only few studies analyzed interactions of SNPs that associated with low HDL-C levels.
In the analysis of genotype data, the effect size of associations between single SNPs and a response of interest are usually small. Thus it is assumed that not only single SNPs but also interactions of several SNP are effective. The logic regression is a novel approach for discovering interactions, specifically Boolean combinations of factors that are associated with the response variable (25). There are several methods proposed in the literature to improve the logic regression such as logic feature selection, (26). It is possible to select important interactions first and to design the final model based upon such predictors. Logic feature selection did not improve the quality of the proposed regression model. However, this method was used as an extension to our regression for low HDL-C level prediction (classification), whose results are shown in the supplementary file, Appendix 1.
Previous studies showed that ApoE gene polymorphisms were associated with CVD and affected the lipid profile. The e4 allele was shown as an independent risk factor for Type 2 diabetes mellitus and cardiovascular disease (27). However, in our study, the relationship between the ApoE careers and HDL-C levels in adolescents was not significant. In our study, ApoE carriers were significantly dependent with rs708272 (P value <0.01), rs2230808 (P value < 0.05), and rs320 (P value < 0.001) polymorphisms. Such variables were significant in our model. Thus, discarding the ApoE variable would decrease the model redundancy. Meanwhile, an association with ApoE and HDL-C, levels has been observed in some but not in all studies in the literature (28, 29).
Our findings showed that individuals without CC genotype of CETP rs708272 polymorphism have upper HDL-C levels. In the other words, T allele of rs708272 polymorphism is associated with raised levels of HDL-C. A meta-analysis published in 2003 has systematically shown that CETP rs708272 polymorphism is associated with HDL-C levels (30). The lack of the T allele of rs708272 polymorphism of the CETP gene was related to CAD in the Chinese population only and T allele of the rs708272 was significantly related to higher HDL-C levels in Chinese male (31). The T allele of rs708272 polymorphism was related to elevated plasma HDL-C levels in the Chinese obese population (32). A national study showed that rs708272 polymorphism was protective on dyslipidemia in Iranian children (33). It was shown that the C allele of rs708272 is related to increased CVD and type 2 diabetes mellitus risks (34). In the other study, the relationship between T allele and higher HDL-C levels in Greek children were observed (35). A meta-analysis of 13,677 individuals indicated that the rs708272 polymorphism was highly related to HDL-C concentration and the risk of atherosclerotic CVD, ultimately (36). Furthermore, a significant relation of the T allele of rs708272 polymorphism with high HDL-C levels has been reported for the Framingham (37), Chinese (38), Iranian (39), and Tunisian populations (40).
We observed that an interaction of rs2230808 polymorphism in ABCA1 gene and rs5880 polymorphism in CETP gene is associated with HDL-C levels. Individuals with GG genotype of rs2230808 polymorphism or G allele of rs5880 have lower HDL-C levels comparing to other subjects indicating that both the main effects of this polymorphism and their interactions can affect the HDL-C concentration. The replacement of C by G at amino acid 373 of CETP resulted in the rs5880 polymorphism. The adverse effect of carriers of the rs5880 polymorphism on the HDL-C levels may be explained by increasing plasma CETP concentration (41). The rs5880, CG, and GG genotypes were affiliated with 17.2%, 95.8% lower large HDL-C particle concentrations, and related with 7% and 41% lower HDL-C levels, respectively (42). The G allele of rs5880 was shown to relate to lower HDL-C, while the ischemic CVD risk was unexpectedly related to being decreased 36% in women with the G allele after HDL adjusting (43). A previous study presented that rs5880 polymorphism was negatively associated with lipid profile and resulted in a 4-fold increase in the childhood dyslipidemia risk (33). The multi-ethnic study of Atherosclerosis (MESA) demonstrated that the G allele of rs5880, which is related to higher CETP concentration (19.5%) and activity (9.4%) and lower HDL-C (6.0%), is also related to atherogenic effects (44). Studies of the ABCA1 gene indicated that its both common and rare variants affect levels of HDL-C and the risk of ischemic CVD (45). It was shown that rs2066718 and rs2230808 were related to HDL-C levels (46, 47). Another study in China showed that G allele of rs2230808 was related to decreased HDL-C level (48). The Russian population found that rs2230808 polymorphism did not affect lipids levels in patients with CVD (49). Additionally, in the population of China, no relationship between rs2230808 with lipids levels in patients with type 2 diabetes mellitus was found (50).
Our result showed that an interaction of rs320 polymorphism in LPL gene and rs708272 polymorphism in CETP gene was associated with HDL-C levels. Individuals with TT genotype of rs320 and TT genotype of rs708272 had higher HDL-C levels. The relationship between the rs320, rs1801177, and rs328 polymorphisms with both TG and HDL-C, as well as myocardial infarction was analyzed in AVCD and control subjects. Moreover, it was observed that carriers of the less common allele of the rs320 polymorphism had 0.04 mmol/L higher HDL-C levels and 0.09 mmol/L lower triglycerides levels than non-carriers (51). The rs320 GG genotype and rs320 G allele were observed to be significantly associated with stroke in Indian population. Also, rs320 GG genotype associated significantly with high levels of TG and low levels of HDL. However, this polymorphism did not show any association with LDL-C and VLDL levels (52). It was reported that in Iranian children and adolescents, carriers of T allele of rs320 polymorphism are associated with lower TG and LDL-C and higher TC and HDL-C (24). Other studies showed that in healthy-weight men with coronary heart disease, the rs320 polymorphism alone might impress the HDL-C concentration, in contrast to rs328 alone, which has no influence on any lipid parameters (53).
Our result showed that an interaction of rs320 polymorphism and rs1801177 polymorphism in in LPL gene was associated with HDL-C levels. Individuals without GG genotype of rs320 or AA genotype of rs1801177 had higher HDL-C levels. The G allele of rs1801177 polymorphism is shown to be associated with lower HDL-C levels and higher LDL-C levels in Iranian adolescents (24). It was reported that rs1801177 polymorphism affects serum lipids, and thus increase the CVD risk (54). However, another case–control study showed that the rs1801177 genotype was not significantly correlated with CVD (55).
4.1. Study Limitations and Strengths
Our study was a cross sectional. This is in fact one of the limitations of our work. However, the pediatric age group and the region under the study are 2 advantages of our work in comparison with the state-of-the art.
4.2. Conclusion:
We showed that rs708272 polymorphism in CETP gene has an important effect on the level of HDL-C, independently. Moreover, rs708272 increased HDL-C levels and had a protective effect on HDL-C. The interaction of ABCA1 (rs2230808) as well as CETP (rs5880) and the interaction of LPL (rs320) as well as CETP (rs708272) were associated with lower HDL-C levels. Furthermore, the interaction of LPL (rs320) and LPL (rs1801177) was associated with lower HDL-C levels.