1. Context
Obesity has become a primary health issue, worldwide, and has stepped into epidemic scales (1, 2). The prevalence of obesity in Iranian adults and children is increasing. Taking into account that childhood obesity increases the risk of adult obesity, these statistics are really menacing for the next generations (3, 4). Obesity has long been recognized as one of the most significant risk factors for multiple nutrition-related non-communicable diseases, including: type 2 diabetes mellitus (T2DM), hypercholesterolemia, hypertension, and heart disease (1, 5). The causes of this deadly epidemic have not been established clearly and fully, yet. However, it is accepted by most researchers and health experts that “obesogenic” environment overall, which comprise the high calorie intake, coupled with the sedentary lifestyle imposed by ‘modern lifestyle’, is a major contributor to this condition (5, 6).
Along with the environment, researchers believe that genetic factors also play an important role in the etiology of obesity, based on the observation that not all individuals with a high calorie intake and low physical activity necessarily get fat (7). Complicated ‘gene-environment’ interactions are mostly responsible for the current “diabesity” epidemic (2, 5, 8-10). Family and twin studies show that 80% of body mass index (BMI) variation, 50% of T2DM risk, and 10‒30% of metabolic syndrome risk are attributable to genetic factors (11). Among factors, affecting genetic variations, there are single nucleotide polymorphisms (SNPs). Although SNPs are not usually the sole disease-inducing factors, they can determine one’s predisposition to special metabolic disturbances and, therefore, to diseases (9).
Among the biological factors playing a role in the etiology of obesity, there are appetite-regulating peptides, including endocannabinoid system (a system consisting of cannabinoid receptors and enzymes involved in the synthesis and degradation of endocannabinoids), ghrelin, neuromedin-beta, leptin, and uncoupling proteins (related to the efficacy of energy metabolism in the human body) (6). Leptin and ghrelin are the two most significant energy- and appetite-regulating peptides.
In 1994, leptin was discovered to be the product of the “obese (ob)” gene and it was named after the Greek word, leptus, meaning thin. Leptin is produced predominantly in adipose tissue and is proportionally related to adiposity. Although leptin levels do fall with fasting and its absence is related to hyperphagia in animals and humans, several of these responses do not necessarily comply with changes in fat mass. This constatation suggests that the most significant role of leptin is in body energy homeostasis and not in meal-to-meal intake (10, 12).
Ghrelin is a naturally-occurring ligand to growth hormone secretagogue receptor (GHSR). Ghrelin is predominantly synthesized in the stomach. In contrast to leptin, ghrelin level in blood rises with hunger and is suppressed by feeding. Again, contrary to leptin, ghrelin is proposed to be more involved in short-term appetite regulation, although it also seems to be a factor in long-term energy homeostasis (12). Clearly, the production and secretion of these peptides and their receptors are regulated by their related genes. Multiple studies have been conducted regarding the effects of the common polymorphisms in these genes and the impact they may exert on weight-related phenotypes. However, since the studies showed inconsistent results and few review articles have been published on this issue, we decided to review the recent papers that dealt with this subject.
The aim of this study was to investigate the association between the most common SNPs in the genes coding leptin and ghrelin and obesity and obesity-related health outcomes. We also expanded our research to SNPs in the genes coding their significant receptors (leptin receptor and GHSR).
2. Evidence Acquisition
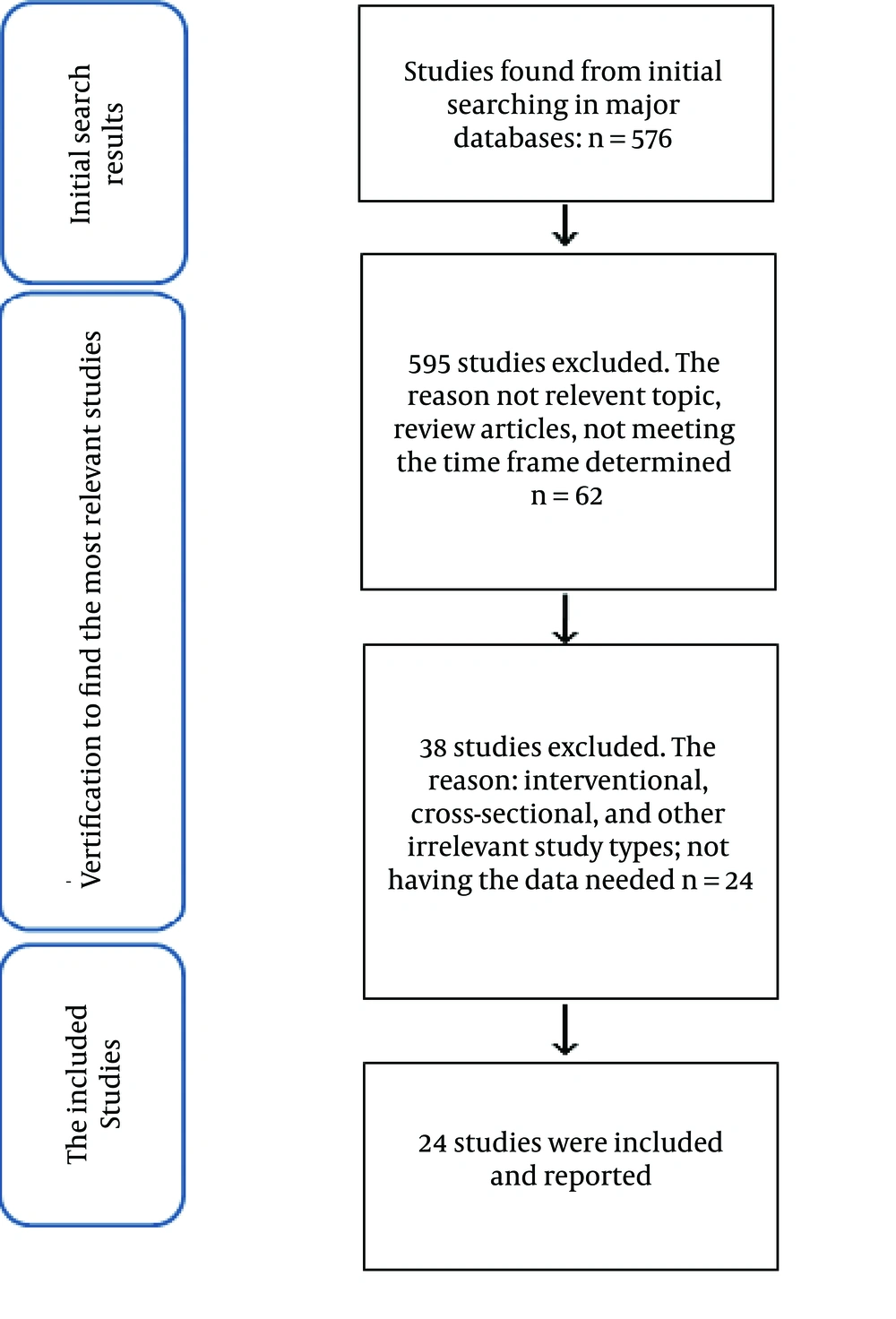
The review of literature was started in November 2013 at the Faculty of Nutrition Sciences and Food Technology, Shahid Beheshti University of Medical Sciences, Tehran, Iran. We used the following keywords to search for the related articles: leptin, ghrelin, polymorphism, single-nucleotide polymorphism/SNP, obesity, overweight, BMI, metabolic disturbances, metabolic syndrome, and T2DM. These keywords were searched in the following websites/databases: Pubmed, Sciencedirect (Elsevier), and Google scholar. The search in databases was made using the following pattern: the exposures (i.e. the SNPs) AND the outcomes (e.g. obesity). The inclusion criteria used to choose the articles were: 1) the relevance of the article to the main title of the review. The studies should at least contain the common SNPs in leptin/leptin receptor genes (LEP/LEPR), ghrelin gene (GHRL), or GHSR and the following outcomes: BMI, waist circumference, adiposity (percentage of body weight, as fat), waist-to-hip ratio (WHR), serum lipid profiles and indicators of glycaemia/T2DM; 2) the articles should match the time frame determined, which includes all the articles published from 2009 to present (just to report more recent studies); 3) the originality of the articles, and 4) only case-control studies were selected to be analyzed. The reason why case-control studies were included in the review was that other kinds of studies were either too small in number (interventional studies) or too insignificant and region-specific (cross-sectional studies) to be report-worthy. Moreover, reviewing studies with the same design gives us the opportunity to draw a logical conclusion, when pooling the results; reporting studies with different study designs would make this much more difficult or even impossible. Exclusion criteria included: 1) animal or cell-culture studies; 2) review studies and case reports, and 3) studies not meeting the inclusion criteria. The review was unrestricted in terms of healthy or obese/diabetic patients, as the cases/controls. Figure 1 shows the diagram according to which the articles were included or excluded from the review.
Finally, the necessary data were extracted from each article. These data included: the details needed for the reference (author, journal, year, etc.), sample size and the relevant characteristics of the cases/controls, the SNPs studied, the phenotypes (obesity-related outcomes), the particular ethnic group studied, if mentioned; the specific allele frequency and the difference between the two groups in this regard and the final conclusion that the authors made about the results of the study.
3. Results
Overall, 657 studies were found and filtered according to our inclusion/exclusion criteria. Out of these, 24 studies matched the criteria and were analyzed further. A total of 633 studies were excluded.
3.1. Study Quality
Firstly, we qualitatively analyzed the quality of the studies, which were included, in terms of sample size, data presentation, and case/control inclusion and exclusion criteria. Regarding sample size, most of the studies used fairly large enough sample size, which would enable to detect the small correlations that are of great importance in genome-related studies. However, the rationale behind determining the sample size was not always justified by the authors. Most of the studies also reported well on subject characteristics (age, gender, weight, BMI, etc.). However, the way the participants were recognized as eligible to take part in the studies (inclusion/exclusion criteria) was not always described. Concerning the obesity-related parameters, most of the studies assessed a number of them.
3.2. Overall Results on Leptin/Leptin Receptor Genes and Ghrelin/Ghrelin Receptor Genes Single-Nucleotide Polymorphisms
As previously mentioned, 24 studies were investigated about common SNPs on LEP/LEPR and GHRL/GHSR and their relation to obesity and its related health parameters. Of these, 17 were on LEP/LEPR SNPs and seven studies on GHRL/GHSR SNPs. The studies on LEP/LEPR were conducted on all continents (nine, eight, four, and three studies were conducted in Asia, Europe, America, and Africa, respectively). Therefore, we can say that this review covers most of the ethnic groups all around the world, making it easier to generalize the results to all healthy people worldwide. However, no studies about GHRL/GHSR were included from America and Africa in this review.
The most prevalent LEP and LEPR SNPs studied were LEP G-2548A and LEPR Q223R, respectively. However, LEPR K109R and K656N were studied more frequently. The GHRL SNP most studied was Leu72Met.
Of the seventeen articles on LEP/LEPR SNPs, nine studies reported significant association with obesity and/or its related health outcomes, or at least considered them as possible risk factors; however, eight of these studies failed to discover such relationships (1, 11-26). Among LEPR SNPs, Q223R was studied in 12 studies. This polymorphism was reported to be related with obesity and/or overweight in four studies (13-16). This SNP was also stated to be a risk factor for T2DM in one study conducted on people with this condition (17).
The K109R and K656N were the other two important SNPs and their association with obesity was investigated in five and four studies, respectively. These two LEPR SNPs were reported to be related to obesity in children, in one study (13), which was not the case in adults. Another study, which studied the relationship between K109R, K656N and LEP 3’ flanking region polymorphism and obesity claimed that these SNPs were associated with obesity only when present simultaneously (18).
It should also be noted that four studies examined the relationship between LEPR SNPs and obesity and/or overweight in children and/or adolescents (13, 19-21). Among these, only Tabassum et al. (13) could find a significant association between Q223R, K109R and K656N and obesity in children.
The most prevalent GHRL SNP studied was Leu72Met. Of the seven studies on GHRL/GHSR SNPs (six on GHRL and one on GHSR), three studies showed significant relationship between these SNPs and obesity and/or its related health outcomes. However, four studies did not show a relationship. All of the studies on the relationship between GHRL SNPs and obesity included Leu72Met (22-28); nevertheless, only two of them (by the same author) reported a significant relationship (22, 26). The Arg51Gln was included in only one study, which found no association (25). Several unpopular GHRL SNPs were also investigated. Surprisingly, these SNPs (including + 3056 T > C (rs2075356), -1500 C > G (rs3755777), -1062 G > C (rs26311), -994 C > T (rs26312)) were reported to be significantly related to obesity (22, 26). One study examined the relationship between GHSR -151 C > T SNP and obesity, which also failed to find a meaningful association (28). Also, it is noteworthy that one study that included children, as participants, could not show a relationship between Leu72Met and obesity in children (24).
A summary of the studies reviewed and their results can be found in Table 1Table 2. The studies are presented in chronological order (from the most recent to the least recent study).
| Authors | Country | Subjects | SNP (rs Number) | Outcome | Reference |
|---|---|---|---|---|---|
| Etemad et al. | Malaysia | 565 people (284 people with T2DM and 281 healthy individuals) | Q223R (rs1137101) | Q223R polymorphism of LEPR might be considered as a significant risk factor for T2DM in Malaysians; especially in Chinese ethnics | (17) |
| Sahin et al. | Turkey | 95 people (47 obese and 48 healthy) | LEP G-2548A | This LEP polymorphism was associated with obesity and serum leptin levels | (29) |
| Lu et al. | China | 432 subjects (230 obese and 202 normal weight) | K109R (1137100) K656N (rs8179183) LEP gene 3’ flanking region polymorphism | These SNPs were significantly associated with BMI only when were present simultaneously | (18) |
| Tabassum et al. | India | 3168 children (2261 normal weight; 907 obese/overweight) | Q223R (rs1137101) K109R (1137100) K656N (rs8179183) | All of these LEPR SNPs were showed to be significantly associated with childhood obesity | (13) |
| Komsu-Ornek et al. | Turkey | 191 children (92 obese and 99 non-obese children) | Q223R (rs1137101) | There was no overall association between Q223R polymorphism of LEPR and obesity and obesity related disorders | (19) |
| Traurig et al. | US | 2842 Pima Indians | 80 different variants were analyzed | The variants rs2025804, rs1171274, and rs6662904 were significantly associated with BMI | (30) |
| Boumaiza et al. | Tunisia | 329 people (160 obese and 169 non-obese) | Q223R (rs1137101) LEP G-2548A | Both variants were significantly related to BMI, energy intake, TC, WC, glycaemia, insulinemia and low HDL-c | (14) |
| Angel-Chavez et al. | Mexico | 128 children and adolescents (76 obese/overweight and 52 normal weight) | Q223R (rs1137101) K109R (1137100) K656N (rs8179183) | There was no association between these SNPs and obesity/overweight in children | (20) |
| Bender et al. | Switzerland | 6184 Caucasians | Q223R (rs10889567) K109R (rs1137100) K656N (rs3790437) | There was no overall association between these LEPR SNPs and overweight/obesity | (1) |
| Angeli et al. | Brazil | 709 people (375 subjects of African ancestry and 334 of European ancestry) | Q223R (rs1137101) LEP A19G | No overall association between these 2 SNPs and obesity/overweight | (31) |
| Pereira et al. | Brazil | 4193 non-diabetic subjects | Q223R (rs1137101) | No special genotype was associated with obesity/overweight | (32) |
| Ben Ali et al. | Tunisia | 710 subjects (393 obese and 317 normal weight) | G3057A LEPR | There was no significant difference between the two groups in genotype frequency | (33) |
| Constantin et al. | Romania | 202 individuals (108 obese and 94 non-obese) | Q223R (rs1137101) LEP G-2548A | There was no overall association between Q223R (in LEPR) and G-2548A (in LEP) with obesity in Romanian subjects | (34) |
| Murugesan et al. | India | 300 subjects (150 diabetic and 150 non-diabetic) | Q223R (rs1137101) K109R (1137100) K656N (rs8179183) | Only Q223R was associated with BMI and significantly different between the two groups | (15) |
| Furusawa et al. | Japan | 809 Pacific Islanders | Q223R(rs1137101) K109R(1137100) LEP G-2548A | Only Q223R was associated to BMI and obesity | (16) |
| Ben Ali et al. | Tunisia | 693 subjects (392 obese and 302 healthy) | Q223R (rs1137101) | There is no overall association between this SNP an obesity | (35) |
| Pyrzak et al. | Poland | 152 children (101 obese and 41 non-obese) | Q223R (rs1137101) | Q223R polymorphism of LEPR is not associated with obesity and other metabolic disturbances in children | (21) |
Summary of Studies on Leptin/Leptin Receptor Genes and Their Association With Overweight/Obesity and Related Health Outcomes a
| Authors | Country | Subjects | SNP (rs Number) | Outcome | Reference |
|---|---|---|---|---|---|
| Takezawa et al. | Japan | 117 obese women | -1500 C > G (rs3755777)1062 G > C (rs26311)-994 C > T (rs26312) Leu72Met (rs696217) + 3056 T > C (rs2075356) | All these variants in ghrelin gene were associated with obesity parameters | (22) |
| Liu et al. | China | 1741 subjects (877 subjects with T2DM and 864 healthy subjects) | Leu72Met (rs696217) | There seems to be no significant difference in Leu72Met distribution between the two groups | (23) |
| Zhu et al. | China | 330 children (230 obese and 100 non-obese children) | Leu72Met (rs696217) | There seems to be no significant difference in Leu72Met distribution between the two groups | (24) |
| Leskela et al. | Finland | 23 MZ twins discordant for obesity (controls); 43 MZ twins concordant for normal BMI and 46 MZ twins concordant for obesity | Leu72Met (rs696217) Arg51Gln | These ghrelin polymorphisms do not affect neither total serum concentration of ghrelin nor the obesity predisposition | (25) |
| Takezawa et al. | Japan | 235 overweight/obese individuals (compared to healthy individuals in HapMap project or other studies analyzing east Asians | -1500 C > G (rs3755777)-1062 G > C (rs26311)-994 C > T (rs26312) Leu72Met (rs696217) + 3056 T > C (rs2075356) | These SNPs are shown to be related to obesity and its related lipidemic and glycemic parameters | (26) |
| Berthold et al. | Germany | 850 Caucasians (420 subjects with T2DM and 430 healthy subjects) | Leu72Met (rs696217) | Leu72Met was associated negatively with T2DM | (27) |
| Gjesing et al. | Denmark | 15854 unrelated, middle-aged individuals | Seven SNPs were investigated; among them: GHSR-151C > T | None of these SNPs were related to quantitative factors of obesity | (28) |
Summary of Studies on Ghrelin/Ghrelin Receptor Genes Single Nucleotide Polymorphisms and Their Association With Overweight/Obesity and Related Health Outcomes a
4. Conclusions
In this article, we reviewed several of the potential SNPs related to energy homeostasis and, consequently, to obesity and the metabolic disorders related to them. These included the genes coding LEP and LEPR and GHRL and GHSR. Of the seventeen articles on LEP/LEPR SNPs, nine studies reported a significant association or at least considered them as possible risk factors; however, eight of these studies failed to discover such a relationship (1, 11-26). Of the seven studies on GHRL/GHSR SNPs (six on GHRL and one on GHSR), three studies showed significant relationship between these SNPs and obesity and/or its related health outcomes, while four other studies did not show a relationship (22-28).
4.1. Overview of Other Studies
Many studies have been conducted on LEP/LEPR and GHRL/GHSR polymorphisms and the effects they may exert on body weight and metabolic disturbances. Since we decided to investigate only the case-control studies published in a particular time period, many of them went unreported in this paper.
Bender et al. conducted a meta-analysis on 55 studies (17 case-control and 38 single group studies) to investigate the relationship between common SNPs of LEPR and overweight (1). These studies were on different races/ethnic groups, including Caucasians, Asians, African, and of mixed ancestry. The most studied SNP, just like in our study, was Q223R. This meta-analysis reported no significant association between LEPR SNPs and overweight, when the data of these studies were pooled; however, several studies, which were analyzed, reported a series of significant associations with markers of overweight/obesity. In another review article by M. Phillips, rs3790433 polymorphism of LEPR was reported to be associated with the risk of metabolic syndrome and insulin resistance (11).
Although we just reported case-control studies, interventional studies are of great importance, when it comes to determining the effects of a gene polymorphism on one’s response to dietary intervention. In a review article by Basic et al., it was reported that a polymorphism in the -2549 position in the promoter region of the LEP gene is associated with obesity and that the carriers of -2549A allele lose less weight in response to a low calorie diet. It was also stated that carriers of the C allele of T343C polymorphism of LEPR lose more weight compared to the non-carriers. Another study in this article reported that Lys656 homozygotes for Lys656Asn polymorphism had a significant loss of body weight, waist circumference, decreased BMI, systolic blood pressure, and leptin levels, in response to a dietary intervention, when compared to carriers of Asn656 (2). Another review article by Rudkowskaand and Perusse investigated the effects of several common LEPR SNPs on the participants’ response to low calorie diets. This article did report the same about the effect T343C polymorphism on weight loss, following a dietary intervention. Another LEP polymorphism, which was investigated in this article, is 3’UTR insertion/deletion; it was reported that the carriers of the I-allele lost greater weight compared to non-carriers. This article also supported that Asn656 allele carriers were more resistant to dietary intervention. Another study reported by this same article showed that Q223R and K109R SNPs of LEPR make people more susceptible to T2DM, although they do not show any relationship with weight loss after a 3-year diet and exercise program (6).
Several review articles also investigated the relationship between GHRL/GHSR polymorphisms and overweight/obesity and related metabolic disturbances. Pulkkinen et al. investigated the most studied SNPs in GHRL/GHSR. Just like our findings, Leu72Met and Arg51Gln were the two most studied SNPs. There have been contradictory results regarding this relationship; several studies proposed that Met72 allele of GHRL was associated with earlier age at onset of obesity and higher BMI, while an inverse relationship has also been reported. This situation resembles the finding of our study. The other SNPs most studied were -501A > C and + 3056T > C; inconsistent results were also reported regarding the effect that these SNPs may exert on body weight. This review also stated that, according to the studies, GHRL variations may have several effects on blood pressure. The GHSR polymorphisms were also shown to be inconsistently associated with features of metabolic syndrome (36). Seven GHSR polymorphisms were analyzed for their role in obesity in the participants of Finnish Diabetes Prevention Study; results show that people with rs490683 C/C genotype were more prone to lose more weight after a 3-year diet and exercise intervention (6).
Taking into account the results of our study and other studies reviewed above, it can be easily understood that giving a sure statement about the association of these SNPs and obesity and its related diseases is no way reasonable. There are several proposed reasons why such contradictory results are seen: 1) the different environmental exposures, which are shown to affect the way genes are expressed (such as nutrient intake) (6), 2) inconsistent methods and criteria used to classify individuals as overweight/obese or lean/normal, 3) unjustified and arbitrary strategies in determining the sample size, and 4) inconsistent genotyping methods, with different precisions, to detect the specific SNP. Besides discovering these associations, there are many controversies that necessitate further studies on this subject: 1) if such a relationship between these SNPs and overweight/obesity really exists; can this specific allele be regarded as an actual risk factor? Answering this question will make researchers and governments determine the prevalence of these SNPs in each and every society. A risk allele will lose its significance when it is not common enough in a particular population, no matter how strongly associated with obesity; 2) what is the real risk SNP, and more specifically, the real risk allele. Even though we reported the most studied SNPs, this does not necessarily mean that they are the actual risk SNPs. Even the real risk allele, in each specific SNP, is not yet fully appreciated; e.g. several studies reported that Met72 is the risk allele in Leu72Met polymorphism of GHRL, while other studies also reported this SNP and allele as protective factors (10, 27, 36). This is also the case in Q223R; contrary to most of the studies, one study reported the Q allele as the risk allele (16).
This study has the following limitations: 1) only studies published between 2009 to the time that this study was conducted were included, 2) the studies included were not filtered regarding their sample sizes, and 3) unfortunately, due to limited number of studies related to each ethnicity, these findings cannot help us discover more about the effect that each SNP may exert in different races and ethnicities.
In this study, we reviewed a total 24 case-control studies, published from 2009 to present. Of these, 17 studies were on LEP/LEPR and seven studies were on GHRL/GHSR and their association with overweight/obesity and/or its related metabolic disturbances. Although several studies show certain relationships, the results were inconclusive and sometimes contradictory. The real association of these SNPs with obesity remains to be clarified and warrants further research and meta-analyses. Our review failed to detect a consistent correlation between these gene polymorphisms and metabolic disturbances, which, in part, may be due to a series of unidentified environmental interactions and possibly the effects that inter-individual variations in nutrient intake might have in gene expression (the nutrigenomics). It should also be noted that case-control studies usually fail to discover clear cause-and-effect relationships and, since we reviewed only case-control studies, this is one of the limitations of our study. Besides case-control studies, a very important amount of work needs to be done in clinical trials to further elucidate the effects of these polymorphisms on different individuals’ responses to dietary interventions.