1. Background
Papillary thyroid cancer (PTC), a follicular epithelial cell-derived thyroid cancer, accounting for approximately 85% of thyroid cancers, is the most common endocrine system malignancy (1). The prevalence of PTC differs in various countries, particularly among Asian populations, in whom it is higher than other ethnicities (2). Although most patients with PTC have a good survival rate, they may be prone to recurrence with increased morbidity and mortality (3, 4).
Existing evidence shows new aspects of molecular mechanisms, such as genetic alterations, which open up a number of research avenues to determine pathways of progression and aggressiveness of PTC. One of the most well known oncogenic genetic alterations, occurring in over 45% of cases of PTC, is the B-type raf proto-oncogene (BRAF) point mutation (5-7). Mutation in BRAF (V600E) is associated with an invasive clinical period and poor prognosis, such as extrathyroidal invasion, lymph node metastasis, and advanced TNM stage (8); the TNM system considers tumor size and local extent (T), lymph node metastases (N), and distant metastases (M). However, the molecular events whereby mutations in the BRAF gene contribute to development and progression of PTC remain unknown.
Recently, regulations in the expression of various tumor suppressor genes that might be affected by mutations in the specific gene have been investigated (9, 10). Hu et al., (10) showed that methylation of various tumor suppressor genes, such as tissue inhibitor of metalloproteinase-3 (TIMP3), may mediate progression and aggressiveness of PTC in response to a mutation in the BRAF gene. TIMP3 is a member of tissue inhibitors of matrix metalloproteinases (MMPs) that play an important role in inhibiting growth, angiogenesis, invasion, and metastasis of various human cancers, including thyroid cancer (11); it is involved in regulation of VEGF-mediated angiogenesis via binding directly to VEGFR2 and preventing the downstream signaling pathways essential for cell stimulation (12). Low levels of TIMP3 expression have been related to the initiation of tumorigenesis, due to decreased anti-MMP, apoptotic activity, and stimulation of angiogenesis. TIMP3 is of particular interest for therapeutic purposes due to its characteristic of inhibiting different aspects of tumor development, mainly because it is a potent angiogenesis inhibitor. Numerous studies on methylation-associated silencing of TIMP3 suggest a tumor suppressor role for it in different cancers (13-17). In addition, hypermethylation of the TIMP3 gene is related to thyroid cancer (18) and correlated with high-risk clinicopathological characteristics of PTC, including extrathyroid invasion, lymph node metastasis, and advanced disease stages (III and IV) (10). However, to the best of our knowledge, no study has so far investigated changes in TIMP3 mRNA and protein levels in papillary thyroid tissues in relation to the BRAF mutation and its clinicopathologic features among PTC patients.
The purpose of this study was primarily to compare the association of TIMP3 mRNA and protein levels with BRAF mutation in PTC patients. In addition, we also aimed to evaluate the association of TIMP3 mRNA and protein levels with the clinicopathologic characteristics in PTC.
2. Materials and Methods
2.1. Patients’ Selection and Data Collection
In the current study, we collected primary PTC tissues (tumoral tissues and adjacent unaffected thyroid tissues for each patient). Clinicopathological data of 60 patients with PTC, who had undergone a total thyroidectomy over a one-year period (2015 - 2016) from the Erfan and Atiyeh hospitals, 2 private hospitals in Tehran city, were enrolled. Clinicopathological data were collected, including information on age, gender, the status of extrathyroidal invasion, lymph node metastasis, multifocality, and the stage of tumor. A pathologist reviewed all samples to confirm the diagnosis of PTC. To determine the tumor stage we used the American Joint Committee on Cancer (AJCC) handbook guidelines (19). Lymph node status was defined as N0 (there was no involvement of nearby lymph nodes), N1a (lymph nodes around the thyroid in the neck were involved), and N1b (other lymph nodes in the neck or lymph nodes behind the throat or in the upper chest were involved). All confirmed thyroid samples were snap-frozen in liquid nitrogen and stored at -80°C.
Ethics approval for the study was obtained from the Ethics Committee of the Research Institute for Endocrine Sciences of the Shahid Beheshti University of Medical Sciences (25ECRIES93/10/23). Written informed consent was obtained from all of the patients.
2.2. DNA Extraction and BRAF Mutation Detection
Genomic DNA was extracted according to the manufacturer’s instructions from the fresh frozen thyroid tissue specimens using the TRIzol reagent (Invitrogen U.S. Cat. No. 15596 - 026), after histological control. Isolated DNA was amplified by the polymerase chain reaction (PCR), carried out using the following specific primers for exon 15 of the BRAF gene containing the site for the T1799A (V600E) mutation: 5’-CTT CAT AAT GCT TGC TCT GAT AGG-3’ (Forward) and 5’- TTC TAG TAA CTC AGC AGC ATC TCA-3’ (Reverse); the PCR reaction was performed in 20μL volume with PCR master mix (Kowsar Biotech Co. Cat. No. K1140), forward and reverse primers (10 pmol), genomic DNA (100 ng/μL), using thermal program including initial denaturation at 95°C for 5 minutes, followed by 40 cycles for denaturation at 95°C for 30 seconds, annealing at 62°C for 30 seconds, and extension at 72°C for 30 seconds. After the last cycle, a final extension was performed at 72°C for 10 minutes. After quality confirmation by agarose gel electrophoresis, direct DNA sequencing of the 251bp PCR-amplified products was carried out, using the same primers by Applied Biosystems 3130/3130xl Genetic Analyzers. The results were analyzed using Chromas software version 2.6.
2.3. RNA Samples and Quantitative Real-Time PCR
Total RNA was extracted from fresh snap-frozen PTC tissue samples after histological control, using the TRIzol reagent (Invitrogen U.S. Cat. No. 15596 - 026), according to the manufacturer’s instructions. RNA quantity and purity were measured using the NanoDrop 1000 (Thermo Scientific, Waltham, and Mass). Total RNA (1μg) was reverse transcribed with the cDNA synthesis kit (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol and stored at -20°C for future use. A amplification of TIMP3 templates was performed by quantitative reverse transcriptase real-time PCR (qRT-PCR) using the Rotor-Gene 6000 (Corbett Research, Sydney, Australia), following thermal program including initial denaturation (10 minutes at 95°C) followed by 40 cycles of 15 sec at 95°C, 20 sec at 60°C, and 40 sec at 72°C. Real-time quantification was monitored by measuring the fluorescence activity. β-actin gene was used as internal control to normalize mRNA levels; all experiments were duplicated. Primers were designed from the sequence of the human cDNAs. Sequences of these primers were as follows: 5’-CTG ACA GGT CGC GTC TAT GA-3’ (Forward) and 5’- CCG ATA GTT CAG CCC CTT GC-3’ (Reverse) for TIMP3, 5’- GAT CAA GAT CAT TGC TCC TCC T-3’ (Forward) and 5’- TAC TCC TGC TTG CTG ATC CA-3’ (Reverse) for β-actin. PCR amplification was performed in 25 µL volumes, using the SYBR Green master mix (Thermo Fisher Scientific, USA).
2.4. Enzyme-Linked Immune Sorbent Assay (ELISA) Analysis of TIMP3 Protein Levels
Total protein was extracted from snap-frozen thyroid tissues. The samples were incised and weighed (totally 100mg tissue/ 1mL buffer); PBS (pH 7.4, 100mM) was added and homogenized thoroughly using a homogenizer (QIAGEN, Germany), and then, centrifuged (at 4000-6000 RPM) for approximately 10 minutes; supernatants were collected and frozen at -20°C for later use. ZellBio GmbH assay kit (Cat. No: ZB-11267-H9648, Germany) was used to quantitatively assay human TIMP3, based on the Biotin double antibody sandwich technology.
2.5. Statistical Analysis
Normal distribution of quantitative data was determined by the Kolmogorov-Smirnov test. Group comparisons of categorical variables were performed using the Chi-square (χ2) test or linear-by-linear association. Categorical variables were expressed as frequency and percentage. For normal distributed data, comparisons of means were evaluated by the independent samples t-test. The intergroup differences were assessed using the one-sample paired t-test (tumoral versus matched non-tumoral tissues). One-way analysis of variance (ANOVA) was used to compare means in 3 groups of lymph node metastasis (N0 to N1b). Correlation between TIMP3 mRNA levels and its protein level was assessed by bivariate (Pearson) correlation analysis. All normal distributed continuous data were expressed as mean ± standard deviation (SD). All analyses were performed using SPSS software (version 20.0 for Windows; SPSS Inc., Chicago, IL). Probability values < 0.05 were used as the cutoff for statistical significance. Relative quantitation of mRNA levels of TIMP3 was performed by the comparative Ct method according to the following formula (20):
∆Ct Tumoral tissues = Ct (TIMP3) - Ct (β actin)
∆Ct Matched non-tumoral tissues = Ct (TIMP3) - Ct (β actin)
∆∆Ct = ∆Ct Tumoral tissues - ∆Ct Matched non-tumoral tissues
Relative expression = 2-∆∆Ct
3. Results
3.1. BRAF Mutation Status in Relation to Clinicopathological Characteristics
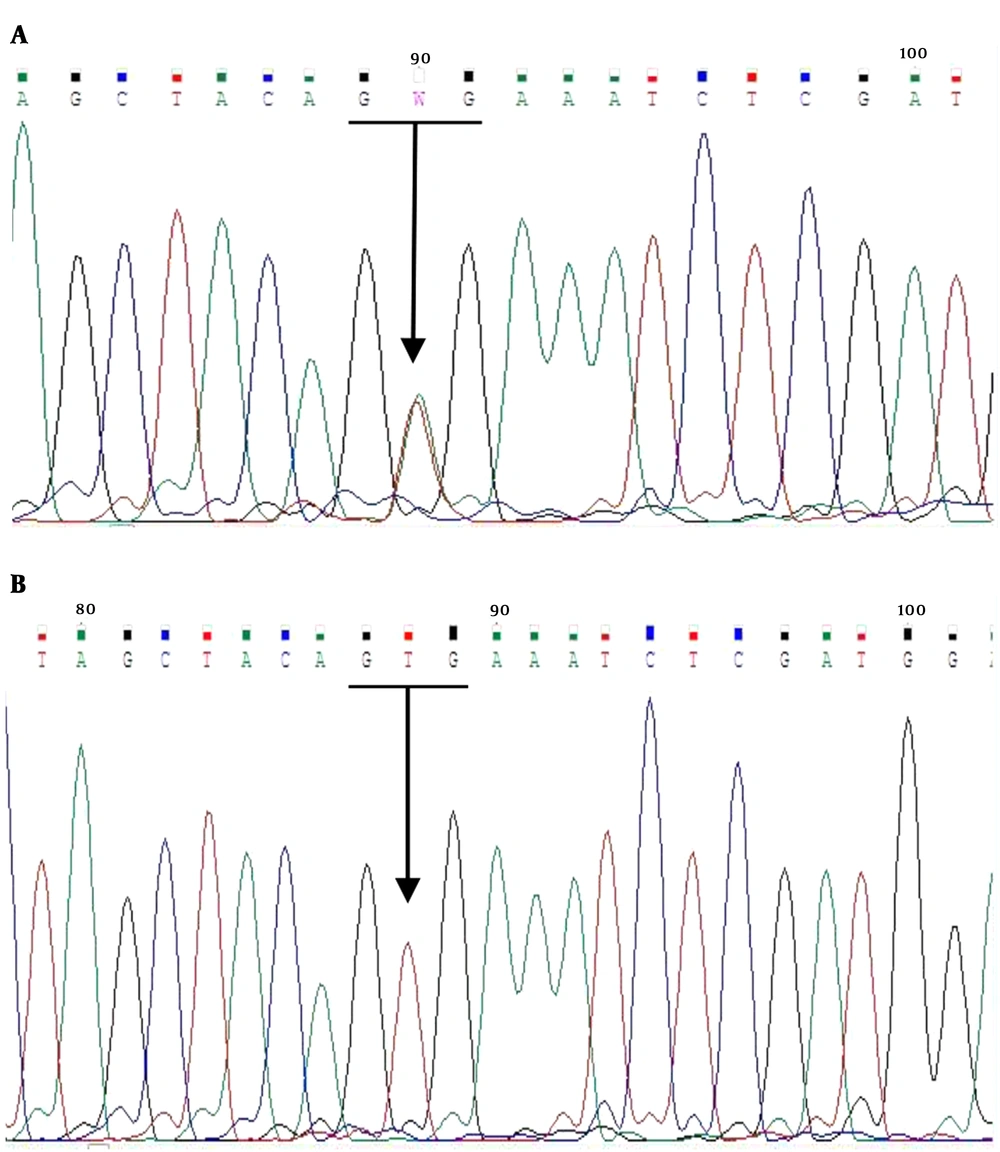
The mean age of males and females was 39.6 ± 15.6 and 36.9 ± 11.7 years, respectively; the BRAF V600E mutation was found in 24 (40%) of the 60 PTC cases. Figure 1 shows a sample of the sequencing results for BRAF (+) and (-). The frequencies of classic, follicular, oncocytic, and sclerosing variants of PTC were 83.3, 11.7, 1.7, and 1.7%, respectively. Using the χ2 test, there was no significant difference in frequency of BRAF mutation in PTC patients (P = 0.322). The prevalence of unifocal and multifocal tumors was respectively 33.3 and 66.7% in BRAF (+) and 38.9 and 61.1% in BRAF (-) (P = 0.534).
We analyzed the clinicopathological characteristics of the PTC cases with BRAF mutation (Table 1). Comparing by independent samples t-test, we found larger tumor size in BRAF (+) PTC than BRAF (-) PTC (mean size 1.98 ± 0.82 vs. 1.31 ± 1.35 cm, respectively; P = 0.039). Lymph node metastasis frequency was significantly higher (χ2 test) in the BRAF (+) compared to the BRAF (-) group (62.5 vs. 34.3%, respectively, P = 0.030).
| Variables | Total | BRAF V600E Mutation | ||
|---|---|---|---|---|
| Positive | Negative | P Value | ||
| Age, years | 40.9 ± 13.7 | 38.8 ± 13.1 | 36.7 ± 12.4 | 0.527 |
| Male, n (%) | 15 (25.0) | 8 (33.3) | 7 (19.4) | 0.180 |
| Female, n (%) | 45 (75.0) | 16 (66.7) | 29 (80.6) | |
| Tumor size (cm) | 1.58 ± 1.20 | 1.98 ± 0.82 | 1.31 ± 1.35 | 0.039b |
| Extracapsular invasion | 14 (23.7) | 6 (25) | 8 (22.9) | 0.544 |
| Lymph node metastasis | 27 (49.1) | 15 (62.5) | 12 (34.3) | 0.030b |
| Lymphovascular invasion | 8 (13.6) | 4 (16.7) | 4 (11.4) | 0.418 |
| Tumor stage (AJCC), n (%) | ||||
| I | 46 (79.3) | 17 (70.8) | 29 (85.3) | 0.153 |
| II | 3 (5.2) | 1 (4.2) | 2 (5.9) | |
| III | 7 (12.1) | 5 (20.8) | 2 (5.9) | |
| IVA | 2 (3.4) | 1 (4.2) | 1 (2.9) | |
Abbreviations: PTC, papillary thyroid carcinoma; AJCC, American Joint Committee on Cancer.
a Data are presented as n(%), except age and tumor size as mean ± SD
b P value < 0.05
| Variable | Tissues Status | Lymph Node Metastasis Status | |||||
|---|---|---|---|---|---|---|---|
| All PTCs | No | Yes | |||||
| No | Yes | BRAF (-) | BRAF (+) | BRAF (-) | BRAF (+) | ||
| TIMP3 protein level, ng/mL | Tumoral | 7.17 ± 3.02 | 7.55 ± 2.63 | 7.47 ± 3.16 | 6.87 ± 3.04 | 8.08 ± 3.39 | 7.08 ± 1.74 |
| Matched non-tumoral | 9.09 ± 3.35 | 12.4 ± 6.78 | 7.58 ± 2.55 | 10.6 ± 3.49 | 12.5 ± 6.84 | 12.2 ± 7.10 | |
| P value | 0.086 | 0.004b | 0.939 | 0.028b | 0.043b | 0.048b | |
Abbreviations: TIMP3, tissue inhibitor of metalloproteinase-3; PTC, papillary thyroid carcinoma.
a Data are presented as Mean ± SD
b P value < 0.05
3.2. TIMP3 mRNA and Protein Levels in PTC
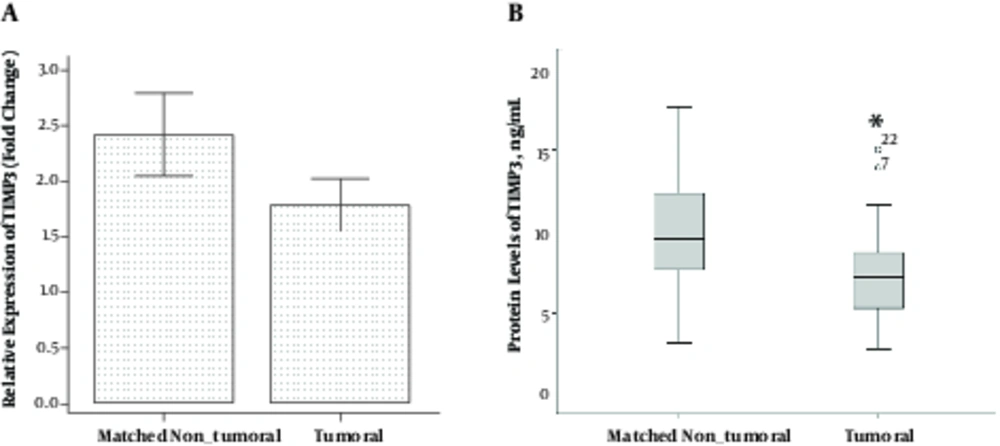
Using one-sample paired t-test, the mRNA levels of TIMP3 did not differ significantly in tumoral tissues compared to those in matched non-tumoral ones (1.79 ± 1.76 vs. 2.42 ± 2.83; P = 0.152) (Figure 2A). The protein levels of TIMP3 was significantly lower in tumoral tissues compared to the matched non-tumoral tissues (7.36 ± 2.79 vs. 10.77 ± 5.57 ng/mL, respectively; P = 0.001) (Figure 2B). Using the bivariate Pearson correlation test, we found no correlation between TIMP3 mRNA and protein levels in tumoral and matched non-tumoral tissues (r = 0.20, P = 0.237 and r = -0.20, P = 0.231, respectively).
Comparison of Tumoral Tissues with Matched Non Tumoral Tissues (Adjacent Unaffected Thyroid Tissues from the Same Patients). A, mRNA Levels of TIMP3 and B, Protein Levels of TIMP3, Error Bars Were Defined as 1 Standard Error of the Mean. Abbreviation: TIMP3, tissue inhibitor of metalloproteinase-3. *P < 0.05.
3.3. Differential mRNA Levels of TIMP3 in BRAF V600E PTC
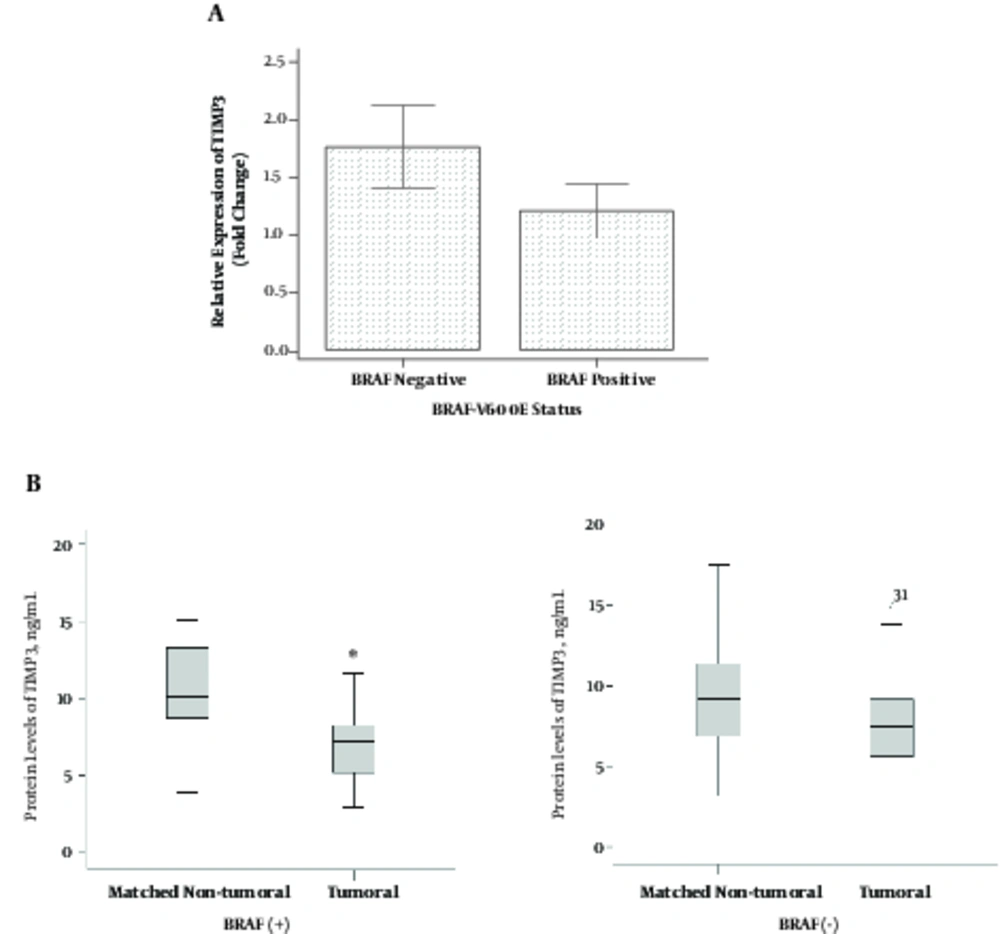
Based on the independent samples t-test, mRNA levels of TIMP3 gene were not significantly different in BRAF (+) PTC compared to BRAF (-) PTC patients (1.21 ± 1.09 vs. 1.76 ± 2.13; P = 0.274) (Figure 3A). Results of the one-sample paired t-test showed that mean of TIMP3 protein level was significantly lower in tumoral tissues compared to the matched non-tumoral tissues in BRAF (+) PTC (6.97 ± 2.37 vs. 11.4 ± 5.59 ng/mL, respectively; P = 0.003), while this level was not significant in BRAF (-) PTC (7.77 ± 3.19 and 10.0 ± 5.61 ng/mL, respectively; P = 0.084) (Figures 3B).
Bivariate Pearson correlation test showed no significant correlation between mRNA levels of TIMP3 and protein levels in tumoral tissues in BRAF (+) (r = 0.19, P = 0.428) and in BRAF (-) PTC (r = 0.19, P = 0.456). No correlation was also seen between mRNA levels of TIMP3 and protein levels in matched non-tumoral tissues in BRAF (+) (r = -0.02, P = 0.925) and BRAF (-) (r = -0.35, P = 0.152).
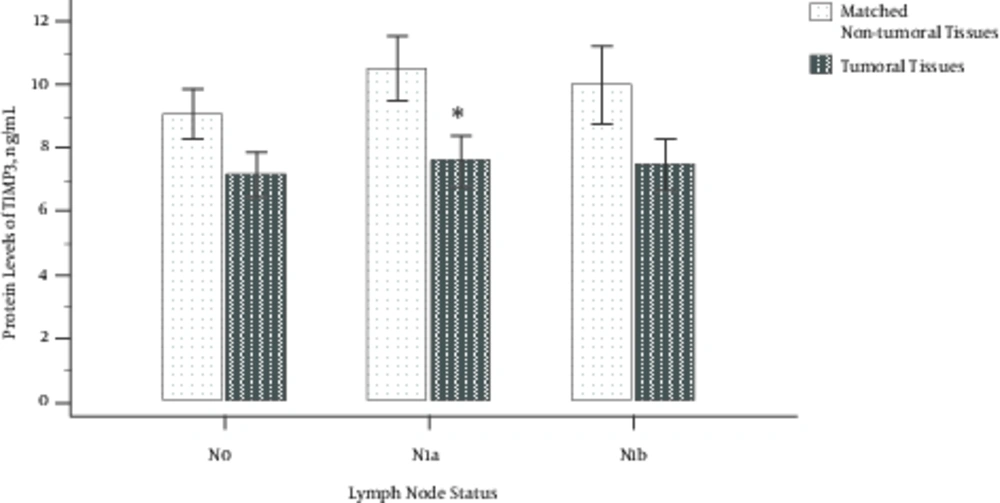
Protein Levels of TIMP3 in Three Different Stages of Lymph Node Metastasis; N0: There Was No Involvement of Nearby Lymph Nodes, N1a: Lymph Nodes Around the Thyroid in the Neck Were Involved, and N1b: Other Lymph Nodes in the Neck or Lymph Nodes Behind the Throat or in the Upper Chest Were Involved; Error Bars Were Defined as 1 Standard Error of the Mean. Abbreviation: TIMP3, tissue inhibitor of metalloproteinase-3. *P < 0.05
3.4. TIMP3 Protein Levels in Different Stages of Lymph Node Metastasis in BRAF V600E (+) PTC
Based on the results of one-sample paired t-test, no association was found between TIMP3 protein levels and clinicopathological features, except lymph node metastasis. In all PTC cases, protein levels of TIMP3 were lower in tumoral tissues compared to the matched non-tumoral tissues of patients with positive lymph node metastasis (7.55 ± 2.63 vs. 12.4 ± 6.78; P = 0.004).
The data file was divided into BRAF positive and negative status. The results using independent samples t-test showed that protein levels of TIMP3 remained significantly lower in tumoral tissues in BRAF (+) in subjects both with and without lymph node metastasis (Table 2).
The protein levels of TIMP3 (using one-sample paired t-test) in tumoral tissues compared to matched non-tumoral tissues in 3 different lymph node status were 7.17 ± 3.02 vs. 9.10 ± 3.34 in the N0 group (P = 0.086); 7.58 ± 2.94 vs. 11.8 ± 5.77 in the N1a group (P = 0.006); and 7.48 ± 2.00 vs. 13.5 ± 9.12 in the N1b group (P = 0.175), respectively (Figure 4). A significant change was observed in the N1a group compared to the N0 but not in the N1b group compared to the N0 one, which may be due to low sample size.
4. Discussion
In this study, we evaluated the most common mutation found in PTC as well as a tumor suppressor gene in tumoral and matched non-tumoral tissues of PTC patients that have not yet been tested. Results showed that the BRAF mutation was related to the tumor size and lymph node metastasis. Moreover, mRNA level of TIMP3 was not significantly changed in tumoral tissue compared to the matched non-tumoral tissue. Furthermore, protein level was downregulated in tumoral tissues in BRAF (+) PTCs; in addition, results indicated that TIMP3 protein level was associated with lymph node metastasis in subjects who was the BRAF V600E (+) mutation.
Data shows that the most frequent genetic variation occurring in PTC is a mutation in nucleotide 1799 of the BRAF gene, giving rise to an activated form, i. e. BRAF V600E (21). Overactivation of the RAF/MEK/MAPK signaling pathway interferes with proliferation, differentiation, and programmed cell death (apoptosis) (22). Furthermore, it has been suggested that the BRAF mutation is associated with a weak prognosis in PTC patients (23). On the other hand, there is increasing evidence suggesting that TIMP3 protects against tumor development by suppressing tumor growth, metastasis, angiogenesis, and apoptosis (24). Reduction in TIMP3 levels in cancer may be due to genetic or epigenetic alterations; genetic mechanisms include changes in gene mutation, polymorphism, and copy number. Su et al., indicated that in patients of oral cancer who carried the TIMP3 polymorphism rs9862 TT genotype had significantly higher TIMP3 plasma levels, compared to the CC genotype (25). Epigenetic mechanisms such as DNA methylation and microRNA regulations can regulate gene expression without changing DNA sequence. In humans suffering from gastric cancer, decreased TIMP3 protein level was correlated with hypermethylation in the promoter region of TIMP3 (26); another study also revealed that TIMP3 protein levels was downregulated by miR-21 in human cholangiocarcinoma (27).
All genetic alternations found in thyroid cancer need to be evaluated for the pathogenesis of this disease, its accurate diagnosis, precise prognosis, and the appropriate management of these patients.
In agreement with our findings, a review by Leonardi et al., (28) of 19 studies, reported a positive correlation between the BRAF V600E mutation with the clinicopathological features, including tumor size, extrathyroidal invasion, lymph node metastasis, and advanced stages in a number of reports on PTC; however, some studies found no such association (29, 30) of an inconsistency, which may be due to various factors, such as different diagnostic criteria and the small number of cases analyzed in different studies.
Similar to our results, Anania et al., (24) presented down regulation of TIMP3 in PTC with respect to normal thyroid tissues using analysis of gene expression data and in silico analysis; they demonstrated that TIMP3 exerts a negative regulatory role in thyroid carcinogenesis. Downregulation of the TIMP3 gene in humans has been demonstrated in melanoma, choriocarcinoma, renal cell carcinoma, esophageal adenocarcinoma, and kidney, brain, and other cancers (16, 31-33). Moreover, TIMP3 promoter hypermethylation is observed in many tumors such as gastric, esophageal, and thyroid carcinoma, and causes decrease in TIMP3 protein levels (26, 34). However, downregulation of TIMP3 protein, not resulting from TIMP3 promoter hypermethylation, has also been reported in pancreatic adenocarcinoma, an occurrence also known to be associated with microRNA and the lack of heterozygosity (15, 35).
In the current study, we found no significant changes in mRNA levels of TIMP3 in tumoral tissues compared to matched non-tumoral ones. However, consistent with a previous study (36), diminished TIMP3 protein level was significantly associated with tumor occurrence. Reduction of protein levels may be dependent on other regulatory mechanisms, which play a role during neoplastic transformations.
Moreover, the protein level of TIMP3 was significantly lower in tumoral tissues compared to the matched non-tumoral tissues in subjects with positive lymph node metastasis in BRAF (+) PTC. No other studies have investigated TIMP3 mRNA, protein levels, and clinicopathological features in relation with BRAF mutation status in PTC, although, Bodnar et al., described that the TIMP3 expression was significantly correlated with lymph node metastases in laryngeal squamous cell carcinoma (37).
The association of TIMP3 protein levels with BRAF mutation status and lymph node metastasis stage confirm the metastasis suppressive function of this protein, which may be influenced by the BRAF gene product. The suppressive effect of V600E mutation on the level of tumor suppressor genes (e.g. TIMP3), which is exerted via MAPK/ERK signaling pathway, has been proposed before (38).
Regarding the study’s strengths to the best of our knowledge, this is the first study being conducted to investigate the association of TIMP3 mRNA and protein levels with BRAF mutation status in tumoral and matched non-tumoral tissues of PTC patients. Other studies have reported only the methylation status of TIMP3 promoter in PTC. However, as for limitations for this cross-sectional study, we had a small sample size and no access to thyroid samples from healthy people (without thyroid carcinoma) to evaluate the probable constitutional changes in TIMP3 levels. Even though we found an association between BRAF mutation and TIMP3 protein level with PTC invasiveness, we did not have a follow-up study to investigate their prognostic role and/or recurrence to give more credibility to the results. The long term diagnosis of BRAF mutation, as the most frequent genetic irregularity in PTC, is not completely understood due to the contradictory findings documented. It is recommended that the prognostic importance of BRAF V600E and low levels of TIMP3 be further evaluated based on long-term outcomes.
In conclusion, our results showed that BRAF mutation was associated with a larger tumor size, higher lymph node metastasis, and lower TIMP3 protein levels. Moreover, lymph node metastasis was seen at lower TIMP3 protein levels.