1. Context
Research of common diseases, currently uses analysis of genetic data available on the functioning of human genome, as the primary most reliable tool for determination of new variants of this genome (1). In addition to genetic variants, environmental and behavioral changes also play an important role in the development of chronic diseases. Therefore, in association studies, gene-environment interactions or presence of a particular environmental exposure, are being strongly emphasized (2). In some situations, populations are related to a lot of exposures in different ways which cause to adjust the effect of specific genetic variants; the resulting interactions can prevent the detection of a genetic effect (3). Collection of pre-morbid exposure can be allowed to standardize by the prospective cohort design. Identifying genomic variations in participants improves the power of these studies. Assessment of environmental risk factors and analyzing gene-environment interactions would provide solid data, less prone to bias and easily extrapolatable to the entire population, compared to case-control studies.
This review highlights some of the important findings documented on genetic cardio metabolic risk factors investigated in the Tehran Lipid and Glucose Study (TLGS) over twenty years.
2. Evidence Acquisition
The search sources for covering a complete range studies were all prominent search engines such as PubMed, Scopus, and Google Scholar with the most proper Medical Subject Headings (MeSH) during the past 20 years.
3. Results
3.1. Project Description
TLGS is a prospective ongoing family based cohort study, conducted on over 15,000 individuals, aged 3 years, enrolled in Phase 1 and followed every 3.5 years since 1999 (4). Other individuals from different pedigrees were also added to the cohort during the follow-up period. Hence, after 20 years of follow-up (2017), 23259 individuals of 5434 households had participated in the study (5). At each visit, subjects signed a consent form and were interviewed to document demographic data including marriage status and spouse information, and were then referred to trained physicians for clinical examinations of the anthropometric and cardiovascular risk factors; they were also referred to laboratories for blood sampling for extraction of sera and DNA. Composition and familial relationship have been described elsewhere (5).
3.2. Heritability and Aggregation
In the present project, the heritability of metabolic syndrome, obesity and lipid profiles of TLGS families were estimated after age and sex adjustment (Table 1) (6, 7). The highest heritability was for HDL-C (about 50%) and the lowest was for metabolic syndrome (15%). Metabolic syndrome components were investigated using exploratory factor analysis (EFA) and results showed that three important factors, including blood pressure, lipids, and obesity in conjunction with fasting blood sugar are the sub components of the metabolic syndrome with heritability of around 6.14 and 7%, respectively (7).
| Cardiometabolic Risk Factors | Mean Heritabilitya, % | P Value |
|---|---|---|
| Lipids | ||
| HDL-C | 0.5 | 8.7 × 10-129 |
| TG | 0.36 | 1.1 × 10-36 |
| FBS | 0.29 | Less than 0.05 |
| SBP | 0.25 | Less than 0.05 |
| DBP | 0.26 | Less than 0.05 |
| LDL | 0.33 | 4.2 × 10-64 |
| Total cholesterol | 0.32 | 1.8 × 10-61 |
| Non-HDL-C | 0.3 | 1.9 × 10-55 |
| Obesity | ||
| BMI | 0.3 | 7.3 × 10-19 |
| WC | 0.27 | Less than 0.05 |
| WHR | 0.27 | 8.0 × 10-17 |
| Waist circumference | 0.32 | 9.1 × 10-22 |
| REE | 0.26 | 4.6 × 10-12 |
| Hip | 0.21 | 1.6 × 10-10 |
| Height | 0.23 | 2.4 × 10-13 |
| Weight | 0.33 | 6.9 × 10-24 |
| Body size | 0.51 | 5.9 × 10-47 |
| High BMI | 0.57 | 5.8 × 10-6 |
| Abdominal obesity | 0.48 | 1.1 × 10-13 |
| High WHR | 0.33 | 0.1 × 10-6 |
| Metabolic syndrome | ||
| Metabolic syndrome score | 0.15 | Less than 0.05 |
Abbreviations: HDL-C, high density lipoprotein; TG, triglycerides; FBS, fasting blood sugar; SBP, systolic blood pressure; DBP, diastolic blood pressure; LDL, low density lipoprotein; BMI, body mass index; WC, waist circumference; WHR, waist to hip ratio; REE, resting energy expenditure.
aHeritability estimation adjusted for age and sex.
Moreover, familial aggregation of the metabolic syndrome components was studied unmarried adolescents who had at least one parent with available information. Results showed a strong correlation between risk of metabolic syndrome in children and parents and significant differences in means of triglycerides, HDL-C and body mass index between children of parents with metabolic syndrome and those whose parents were metabolic syndrome free (8).
3.3. Genetic Association Studies
Genetic association studies, powerful tools used in investigating the correlation between disease status and genetic variation, are helpful in identifying candidate genes or genomic regions that contribute to a specific complex disorder (9). Usually, the candidate genomic regions for different studies are selected based on the results of previous studies, especially previous genome wide association studies conducted on the same population or on other similar populations. Here, we described the most important genetic findings of studies conducted on TLGS participants since 1999.
3.4. Lipid Metabolism
Low HDL-C concentration is the most frequent lipid related risk factor in Iran (10). Some missense mutations in ABCA1 (ATP binding cassette subfamily a member 1), APOA1 (apolipoprotein A1), APOE (apolipoprotein E), APOA5 (apolipoprotein A5), and SCARB1 (scavenger receptor B1) showed significant associations with decrease of HDL-C (11-18); other HDL-C related variations were observed in the splice site, intronic, upstream and downstream regions. The most studied intronic variations in the CETP gene in different populations are rs753562256 (TaqI) and rs708272 (-629A/C) (19). In TLGS participants, allele frequencies were 0.382 for the Taql B2 and 0.462 for the -629A/C A. In addition, linkage disequilibrium analysis showed that Taql B1 and B2 alleles and the alleles of the-629A/C variant were in linkage disequilibrium in our population (D = 0.0965 and 0.4695, respectively). There was a significant association between the presence of B2 and A alleles with higher HDL-C concentration and low CETP activity (20, 21).
In 2012, the effects of two polymorphisms, rs63750792 and rs2070665 were studied in the TLGS population and significant associations were found between triglyceride, HDL-C, HDL2, and levels of Apo AI and Apo B (14). Some years later, Bandarian et al. in an exon and promoter sequence analysis conducted among 63 individuals according to 95th percentile of HDL-C (extremely high) on apolipoprotein A1; their sequencing results showed 42 common and rare variants. Exonic variants included eleven missense, six synonymous, and one nonsense (13).
Regarding the huge effect of the APO gene cluster on lipid profile measurements and the large number of genes in this family, XbaI Apo lipoprotein B polymorphisms were found to have a significant association with increased total cholesterol and Apo lipoprotein B in a cross-sectional study of 849 TLGS participants, associations that remained significant even after adjusting for demographic and blood covariates i.e., age, sex, body mass index, smoking, diastolic and systolic blood pressure, and fasting blood sugar (22).
Mutations in the LDL-R (low density lipoprotein receptor) gene could lead to autosomal dominant disorder, familial hypercholesterolemia. In this gene, the splice region variant rs72658867-A and rs17248748-T in intron 14 and intron 1 are associated with lower non-HDL-C levels and confer protection against CAD. Retention of intron 14 during transcription is caused by the LDLR splice region variant, rs72658867-A, located at position +5 in intron 14 (NM-000527: c.2140 + 5G > A) and can appear a truncated LDL receptor lead to mainly lacked functions of the receptor. This retention of the intron characterizes almost half of the transcripts generated from chromosomes carrying rs72658867-A. Also, in spite of the wild type transcripts do not exceed levels in non-carriers the same variant increases LDLR mRNA expression and this demonstrated that the sequence variants disrupt the LDL receptor can lower non-HDL-C and protect against CAD (23).
One of the well-known variations related to HDL-C concentration is rs1800588 (-514C/T), which may influence HDL-C (24); generally, the (T) allele is considered to lead to higher HDL-C levels. The “T” allele non-carriers in the -514C/T polymorphism of the LIPC gene have lower level of HDL-C than carriers.
SRBI or SCARB1 (scavenger receptor class B member 1) is a key component in reverse cholesterol transportation. In a study conducted to assess the association between lipid profiles and rs4238001 polymorphism through the SR-BI gene among the TLGS residents, minor allele frequency of the polymorphism of SR-BI gene was 0.159 (18); in addition, the association of rs4238001 with HDL-C and HDL-3 was not significant except after adjusting for age. The result of this study showed that age has a confounder role which could regulate the strength of association between the single nucleotide polymorphism of SR-BI and HDL-C level trait (18).
Relationships of the LCAT (lecithin-cholesterol acyltransferase) gene with lipid profiles have also been investigated. Through this study, the promoter, coding regions and exon/intron boundaries of LCAT were amplified. As a result, sequenced in consecutive individuals (n = 150) who had hardly low or high HDL-C levels with no other major lipid abnormalities; 14 single-nucleotide polymorphisms (SNPs) were determined included 10 novel SNPs. Three of the novel variations (position 5,151 in exon 1, position 6,531 in exon 5, and position 6,696 in exon 5) are caused by an amino acid substitution. However, results did not show very strong associations between recognized variants and HDL-C (25). Also, a replicate study conducted on rs5923 polymorphism of the LCAT gene showed no associations with low HDL-C levels in an Iranian population (26).
3.5. Metabolic Syndrome
Genetic studies of the metabolic syndrome in the TLGS, use one of the three following definitions: (1) “National Cholesterol Education Program Adult Treatment Panel III (ATP III)” (27), (2) International Diabetes Federation (IDF) (28) and (3) The Joint Interim Statement (JIS) (29). Furthermore, based on national reports, Azizi et al. suggested a waist circumference cutoff ≥ 95 cm for those aged above 16 years, and WC > 78 cm for those aged between 10 - 16 year (30).
Association studies of genetic markers in the APO gene cluster regions with dietary patterns conducted on patients with metabolic syndrome showed that the rs5128 is associated with different nutrient components in metabolic syndrome affected individuals (31, 32).
Moreover, the relation of variation rs5110 (G360T) with metabolic syndrome has been evaluated in a case/control study (16). Results showed that APOA5 rs2075291 could play an important role in triglyceride and HDL-C levels in individuals affected with metabolic syndrome, although the association of APOA5 rs662799 polymorphism with these levels is still under debate (33).
CD36 (CD36 Molecule), MC4R (melanocortin 4 receptor), and SLC30A8 (solute carrier family 30 member 8 (ZnT-8)) are other important genes that have been reported to be associated with metabolic syndrome in TLGS families (34).
3.6. Obesity
CVD risk factors of obesity have a high heritability as shown in Table 1. Many loci have been reported to be associated with obesity risk factors worldwide. Obesity related genetic regions focused on the effect of the 16q12.2 region, which contains the FTO (fat mass and obesity-associated protein), IRX (iroquois-class homeodomain protein), and MMP (matrix metallopeptidase) family genes, FABP2 (fatty acid binding protein 2), ADRB3 (adrenoceptor beta 3), IL6 (interleukin 6), PPAR-γ (peroxisome proliferator-activated receptor gamma), TNF (tumor necrosis factor-alpha), and CCND2 (cyclin D2) genes.
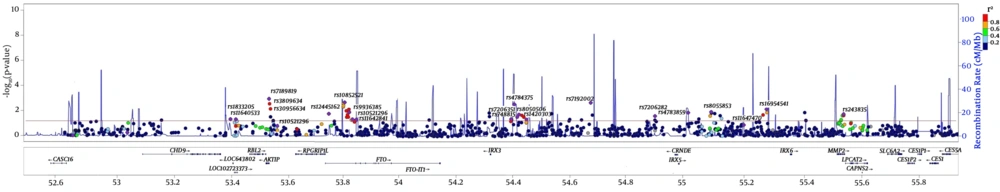
The chromosomal region 16q12.2 contains the most obesity related genes. Various GWA and Meta GWA have provided clear evidences confirmed that FTO is associated with body mass index, and affects dietary intakes and preferences for certain energy-dense foods, as well (35) (Figure 1). In a study conducted on 6928 unrelated subjects with 986 genotyped SNPs located on the 16q12.2 regions, significant SNP sets were relatively denser in the introns of FTO, AIKTIP, and MMP2 genes and near IRX3 gene (Figure 1) (35). Although previous studies confirm the association between rs1421085, rs1121980, rs1558902, and rs8050136 in the first intron of the FTO gene and body mass index, this review showed no association between FTO variants and body mass index in healthy metabolic obese (HMO) individuals (36). FABP2 genes are thought to play a role in the intracellular transport of long-chain fatty acids and their Acyl-CoA esters and may help in maintaining energy homeostasis by functioning as a lipid sensor. Examining a missense variant (rs1799883) on the FABP2 gene among individuals with body mass index ≥ 30 Kg/m2 showed no significant differences between case and control groups in terms of allele frequency (37).
Regional association plot shows the P values (-log P values) of each SNP and recombination rate in the 16q12.2 region. The red line is related to -Log (0.05), indicating significance level. For each significant set, SNP with the lowest P value is determined (35).
In the ADRB3 gene, an association was found between variations in rs4994 and obesity in TLGS population (38). A case-control study examining the relation of rs1801282 polymorphism in the PPAR-γ gene with obesity in 479 participants the TLGS indicated that the presence of G allele could lead to a 1.7 fold increase in the risk of obesity (39).
Regarding previous studies, elevated TNFα levels are linked to obesity and insulin resistance in some populations; the association of obesity with rs1800629 and rs361525 (in the promotor region) was investigated in an adult Iranian population and results showed that these SNPs were not an important risk factor for obesity or consequently for cardiovascular disease (40).
CCND2 encodes a G1/S cell cycle regulator and is the key regulator of postnatal pancreatic β cell mass (41). This cyclin forms a complex with CDK4 or CDK6 and functions as a regulatory subunit of the complex, whose activity is required for cell cycle G1/S transition. In the TLGS population, a variant with low frequency of 1.47% in intron 1 of CCND2 gene, rs76895963, showed significant associations with both increased height and body mass index showed significant associations with both increased height (1.17 cm per allele, P = 5.5 × 10-12) and body mass index (0.56 kg/m2 per allele, P = 6.5 × 10-7). Moreover, the G allele of this variant reduces risk of type II diabetes by 50% (odds ratio (OR) = 0.53 (P = 5.0 × 10-21)) (42).
3.7. Diabetes
In accordance with the definition provided by the American Diabetes Association, adult participants are considered to have diabetes if they meet at least one of the following criteria: FPG ≥ 126 mg/dL, or 2h-PCPG ≥ 200 mg/dL or taking anti-diabetic medication (43). In addition to TCF7L2 (transcription factor 7 like 2) and HHEX, which encode for proteins implicated in blood glucose homeostasis, the effects of PDX1 (pancreatic and duodenal home box 1) and PAM (pancreatic and duodenal homeobox1) genes on type II diabetes (T2D) in TLGS families were investigated.
The PAM gene encodes a multifunctional protein; two missense variants in this gene carrying p.Asp563Gly (with frequency of 4.98% and OR = 1.23, P = 3.9 × 10-10) and p.Ser539Trp (with frequency of 0.65%, OR = 1.47, P = 1.7 × 10-5), allow moderately higher risk of T2D (42).
A transcriptional activator of several genes account for insulin, somatostatin, glucokinase, islet amyloid polypeptide, and glucose transporter type 2 is encoded a protein by PDX1. In our population a rare (0.20%) frameshift variant in PDX1, carrying p.Gly218Alafs*12, demonstrated an association with high risk of T2D (OR = 2.27, P = 7.3 × 10-7) (42).
3.8. Thyroid
Previous studies confirm our findings on the association between TPO (thyroid peroxidase) gene polymorphisms with both anti-TPO and anti-TG factors. TPO or iodide peroxidase is a key enzyme expressed in the thyroid for hormone foundation. TPO catalyzes oxidation of iodide ions to iodine atoms that form iodothyronines in a thyroglobulin (Tg) molecule for the production of thyroxine (T4) or triiodothyronine (T3) hormones (44). Serum autoantibodies in patients with Graves’ disease or Hashimoto’s thyroiditis recognized the TPO that is the major thyroid autoantigen (45). The TPO is a membrane-bound glycoprotein and which in humans, consists of 933 amino acids that are encoded by the mRNA of 3048 nucleotides. The TPO gene consists of 17 exons and extends over 150 kb on the short arm of chromosome 2, locus 2p25. According to molecular genetic studies, mutations in this gene are associated with one of the most common causes of autoimmune thyroid diseases i.e. autosomal recessive inheritance (46, 47). Autoimmune thyroid diseases result from the interaction of several mechanisms, including total absence of TPO activity, inability of TPO to bind to the hem cofactor, inability to interact with the Tg substrate, and abnormal sub cellular localization, all of which are affected by various mutations in the TPO gene. Of TPO gene variations, the associations of C2145T (rs732608), G1193C (rs2175977), and A1936G (rs10189135) polymorphisms with anti-TPO levels were evaluated in the TLGS population. Results showed that the C allele polymorphism in the synonymous variant C2145T of exon 12 is associated with high levels of serum anti-TPO and that carriers of this allele are predisposed to autoimmune thyroid disease 9.2 fold higher than those, who have no C allele. Moreover, missense variant G1193C of exon 8 has no effect on increased levels of anti-TPO (48), although, in the missense variant A1936G of exon 11, the G allele is significantly associated with increased anti-TPO levels (49); all the above results suggest that the rs732608 and rs10189135 polymorphisms are associated with high levels of anti-TPO in this population (49).
3.9. Polycystic Ovary Syndrome (PCOS)
The most prevalent endocrinopathy in female is PCOS. This disorder affects 6% - 10% of caucasian premenopausal women (50), with a reported prevalence of 7% among Iranian women, based on the NIH definition (51).
Associations between PCOS and variations in three different genes, TCF7L2, HHEX, and VDR (vitamin D receptor) have been investigated. TCF and HHEX genes have been implicated in blood glucose homeostasis and genetic variants of this gene are associated with increased risk of T2D. Given the simultaneous occurrence of PCOS and T2D (indicating a common underlying genetic etiology) (52), the association between IR and two strongly T2D associated gene variants (rs7903146 in TCF7L2, and rs1111875 in HHEX) is affected by PCOS status in Iranian women. After adjustment for age and body mass index, a protective effect of the HHEX A allele is observed on the risk of IR in non-PCOS subjects (confidence interval (95%): 0.33 - 0.78; P = 0.002) (52).
The VDR gene encodes the nuclear hormone receptor for vitamin D3. A single nucleotide polymorphism rs757343 in the initiation codon were studied (53-55) and the results showed that presence of the A allele was associated with a 74% increased risk of severe phenotype development (OR, 1.74; 95% CI, 1.07 - 2.82) while no association was observed with disease risk.
3.10. Hip Osteoarthritis and CHADL Gene Variant
The CHADL (chondroadherin like) in the 22q13.2 region is a negative modulator of the chondrocyte differentiation. In an Iranian population the allele frequency of a frameshift mutation, rs532464664 (p. Val330Glyfs*106) was 0.0115 and 23 of the studied individuals were heterozygous for this variation (56).
3.11. Tehran Cardio Metabolic Genetic Study (TCGS)
Considering the importance of genome wide association studies, exome sequencing and whole genome analyses, TCGS was designed in collaboration with the Research Institute for Endocrine Sciences (RIES) and the deCODE genetics company. The TCGS cohort comprises 17,186 (86.3%) of the 19,905 TLGS participants who provided baseline blood samples for plasma and DNA analysis. This study comprises of 849 independent individuals and 3109 families with at least one member with complete genotype data. Studies took account into the TCGS have been being navigated to determine relevant specimen of genetic polymorphisms that could be associated to cardio-metabolic risk factors in Tehran. TLGS database, the oldest and biggest Iran cohort, includes clinical, behavioral, and biochemical data for the whole participants. After augmenting this study by starting TCGS project and appending genome-wide data to the TCGS database, the study would be capable to consider gene-gene and gene-environment interactions and their relation to disease status (5).
4. Conclusions
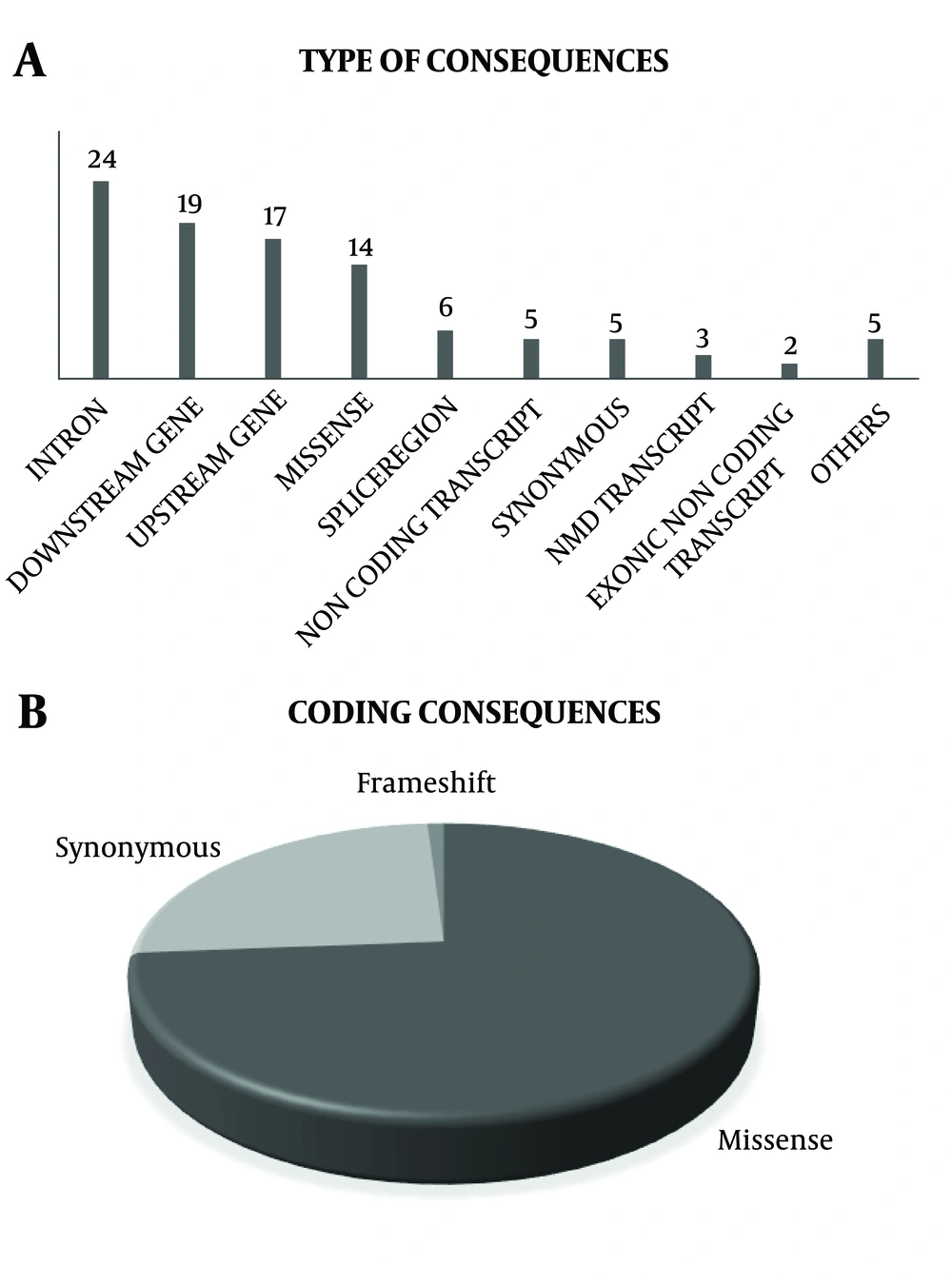
Iran is a large country with diverse ethnicities, with a high rate of consanguineous marriage in some regions resulting in high inbreeding rates and different ethnicities and complex genetic pattern for this population. Considering that, Tehran, the capital city of Iran, has diverse ethnicities and cultures, the TLGS population could provide us with valuable information on Iranian populations. Since, these individuals have been followed for at least 20 years, their phenotype and genotype variations, in addition to their outcomes provide us with good infrastructure for research, the findings of which could shed more light on non-communicable disorders. A summary of genetic association studies has been shown in Table 2 and Figure 2A and 2B. Totally 48 variations in 26 genes in relation to the phenotypes were studied in the TLGS. Among them, 34% of studied variants were in coding consequences, and 20 of them were missense variants. Currently, all participants of the TCGS are chip-typed and over 1000 individuals have been genome sequenced. Data of the TLGS and related studies not only add to current knowledge on genomic differences between Iranian populations and other countries but also recommend new strategies for early prevention and treatment of non-communicable diseases. Moreover, results of the TLGS delineate the path to personalized medicine and predictive medicine, which benefit both individuals and governments providing cost-effective treatments and increasing the quality of life especially in elderly patients.
| Symbols | SNP | Location (Allele) | Consequence | Codons (Amino acids) | Studied Phenotypes/Traits | Sample Size | References |
|---|---|---|---|---|---|---|---|
| ABCA1 | rs2230806 | 9: 104858586 (T) | Missense | aGg/aAg (R/K) | Cholesterol, triglyceride, HDL-C, apolipoprotein A1, apolipoprotein B | 823, 778 | (11) |
| ADRB3 | rs4994 | 8: 37966280 (G) | Missense | Tgg/Cgg (W/R) | Obesity, BMI | 401 | (38) |
| APOA1 | |||||||
| rs2070665 | 11: 116836968 (G) | Downstream | - | Extreme high and low HDL | 132 | (12, 13) | |
| rs121912724 | 11: 116836361 (C) | Missense | cTg/cGg (L/R) | Extreme high and low HDL | 132 | (12, 13) | |
| rs201148448 | 11: 116837080 (A) | Missense | Gtg/Ttg (V/L) | Extreme high and low HDL | 132 | (12, 13) | |
| APOA2 | |||||||
| rs6413453 | 1: 161222526 (A) | Splice region, Intron | - | Extreme high and low HDL | 132 | (12, 13) | |
| rs5069 | 11: 116837538 (A) | Intron | - | Cholesterol, triglyceride, HDL-C, apolipoprotein A1, dietary pattern | 823, 828 | (32, 57) | |
| rs670 | 11: 116837697 (T) | 5-prime-UTR | - | Cholesterol, triglyceride, HDL-C, apolipoprotein A1, dietary pattern | 823, 828 | (32, 57) | |
| rs5082 | 1: 161223893 (A) | Upstream | - | Extreme high and low HDL | 132 | (12, 13) | |
| APOA3 | rs5128 | 11: 116832924 (C) | Downstream | - | Cholesterol, triglycerides, HDL-C, apolipoprotein A1, Dietary fatty acids, dietary pattern, metabolic syndrome | 823, 1510, 828 | (32) |
| APOA5 | |||||||
| rs662799 | 11: 116792991 (A) | Upstream | - | Metabolic syndrome, HDL-C | 947 | (16, 58) | |
| rs2075291 | 11: 116790676 (A) | Missense | Ggc/Tgc (G/C) | Metabolic syndrome, HDL-C | 947 | (16, 58) | |
| rs3135506 | 11: 116791691 (A) | Missense | tCg/tTg (S/L) | Metabolic syndrome, HDL-C | 947 | (16, 58) | |
| APOB | rs693 | 2: 21009323 (A) | Synonymous | acC/acT (T) | Cholesterol, triglyceride, HDL-C, apolipoprotein B | 849 | (14) |
| APOE | |||||||
| rs7412 | 19: 44908822 (T) | Missense | Cgc/Tgc (R/C) | HDL-C, LDL-C, obesity, BMI | 1030, 843 | (17, 59) | |
| rs429358 | 19: 44908684 (C) | Missense | Tgc/Cgc (C/R) | HDL-C, LDL-C, obesity, BMI | 1030, 843 | (17, 59) | |
| CCND2 | rs76895963 | 12: 4275678 (G) | intron | - | Diabetes, height, BMI | 1,624 cases, 9,163 controls | (42) |
| CD36 | |||||||
| rs10499859 | 7: 80629494 (G) | Intron | - | Metabolic syndrome, HDL-C | 337 | (34, 60) | |
| rs13246513 | 7: 80677435 (G) | Downstream | - | Metabolic syndrome, HDL-C | 337 | (34, 60) | |
| CETP | |||||||
| rs708272 | 16: 56962376 (A) | intron | - | Cholesterol, triglycerides, HDL-C | 1021, 555 | (20, 21) | |
| rs1864163 | 16: 56963321 (A) | Intron | - | Cholesterol, triglycerides, HDL | 1021, 555 | (20, 21) | |
| CHADL | rs532464664 | 22: 41238083 (insCGCGCGCC) | Frameshift mutation | gtg/gGGCGCGCGtg (V/GRAX) | Hip osteoarthritis | 996 | (56) |
| FABP2 | rs1799883 | 4: 119320747 (A) | Missense | Act/Tct (T/S) | Obesity, BMI | 400 | (61) |
| FTO | |||||||
| rs1421085 | 16: 53767042 | Intron | - | BMI, MUHO | 945 | (62) | |
| rs1558902 | 16: 53769662 | Intron | - | BMI, MUHO | 945 | (62) | |
| rs1121980 | 16: 53775335 | Intron | - | BMI, MUHO | 945 | (62) | |
| rs8050136 | 16: 53782363 | Intron | - | BMI, MUHO | 945 | (62) | |
| HHEX | rs1111875 | 10: 92703125 (T) | Intergenic | - | PCOS | 504 | (52) |
| IL6 | rs1800795 | 7: 22727026 (G) | Intron | - | Obesity, BMI | 214 | (63) |
| LCAT | rs5923 | 16: 67940050 (A) | Synonymous | Ctg/Ttg (L) | HDL-C | 130 | (64) |
| LDLR | |||||||
| rs200238879 | 19: 11105602 (C) | Splice region, intron | - | Non-HDL, triglycerides, HDL-C | 9,631 | (23) | |
| rs17248720 | 19: 11087511 (T) | Upstream | - | Non-HDL, triglycerides, HDL-C | 9,631 | (23) | |
| rs17248748 | 19: 11095364 (T) | Intron | - | Non-HDL, triglycerides, HDL-C | 9,631 | (23) | |
| rs72658867 | 19: 11120527 (A) | Splice region, intron | - | Non-HDL, triglycerides, HDL-C | 9,631 | (23) | |
| LIPC | rs1800588 | 15: 58431476 (T) | Upstream | - | Cholesterol, triglycerides, HDL-C | 1021, 555 | (65) |
| MC4R | rs12970134 | 18: 60217517 (A) | Intergenic | - | Dietary pattern, metabolic syndrome | 815 | (66, 67) |
| PAM | |||||||
| rs35658696 | 5: 103003107 (G) | Missense variant | gAt/gGt (D/G) | Diabetes | 1,624 cases, 9,163 controls | (42) | |
| rs760687925 | 1: 204973276 (G) | Missense, splice region | tCg/tGg (S/W) | Diabetes | 1,624 cases, 9,163 controls | (42) | |
| PPAR-γ | rs1801282 | 3: 12351626 (G) | Missense | Cca/Gca (P/A) | Obesity, BMI | 239 | |
| SCARB1 | rs4238001 | 12: 124863717 (T) | Missense | Ggc/Agc (G/S) | Cholesterol, triglycerides, HDL | 774 | |
| SLC30A8 | rs13266634 | 8: 117172544 (T) | Missense | Cgg/Tgg (R/W) | Dietary patterns | 816 case and 816 control | |
| TCF7L2 | rs7903146 | 10: 112998590 (T) | Intron | - | Diabetes | 11000 | |
| TNF | |||||||
| rs1800629 | 6: 31575254 (A) | Upstream | - | Obesity, BMI | 244 | ||
| rs361525 | 6: 31575324 (A) | Upstream | - | Obesity, BMI | 244 | ||
| TPO | |||||||
| rs732608 | 2: 1496127 (C > T) | Synonymous variant | (Pro715) | Weight, Anti- TPO, Anti-Tg | 184 | ||
| rs2175977 | 2: 1477459 (G > C) | Missense variant | (S/T) | Weight, Anti- TPO, Anti-Tg | 184 | ||
| rs10189135 | 2: 1493885 (A > G) | Missense variant | (V/M) | Anti TPO, weight | 190 | ||
| VDR | |||||||
| rs757343 | 12: 47845892 (T) | Intron | - | PCOS | 260 | ||
| rs1544410 | 12: 47846052 (T) | Intron | - | PCOS | 260 |
A, All genetic variation consequences studied in TLGS; B, coding consequences studied in TLGS (35)