1. Background
Campylobacter is a zoonotic bacterium frequently associated with acute gastroenteritis in both developing and industrialized countries. Human campylobacteriosis is predominantly caused by Campylobacter jejuni (C. jejuni) and Campylobacter coli (C. coli), although C. jejuni is responsible for the majority of these infections (1). The disease presentation can vary from a mild watery diarrhea to bloody dysentery as the organism colonizes the small intestine of the human host early in infection and later moves to the colon, which is the target organ (2). Campylobacter jejuni is also frequently associated with the development of immunoreactive complications such as polyarthralgia, Guillain- Barre and Miller Fisher syndromes and eventually death (3).
The cause of these discrepant outcomes of infection in humans is not clear although several studies have suggested that various virulence factors present in campylobacters contribute to their survival and the establishment of disease in the host. Motility, bacterial adhesion, invasion of the intestinal epithelium and production of toxin and hemolysin appear to be the main virulence factors. Bacterial adhesion and invasion are well-established early events before the initiation of the inflammatory processes and diarrheal development (4).
The invasiveness of C. jejuni strains plays a vital role in the pathogenesis of this organism and is often used as a measure of bacterial virulence, reflecting the involvement of multiple bacterial structures and mechanisms in this process. Although these pathogens are generally considered invasive, the level of invasion of intestinal epithelial cells in vitro varies among strains (5).
During the initial stage, Campylobacter adheres to the human intestinal cell lining and then is internalized within the cells causing tissue damage, inflammation and thereby gastroenteritis. The bacterial factors implicated in host cell invasion are capsular polysaccharide (CPS), flagella, sialylation of the lipooligosaccharides (LOS) outer core or Campylobacter invasive antigens (Cia) (6-8). It is well known that the polar flagellum present in C. jejuni is crucial for the initial interactions of this organism with its host and facilitates the colonization of the intestinal epithelial cells (9, 10). However, the precise invasive manner of C. jejuni in the disease pathogenesis in humans still needs further clarification.
Moreover, bacterial toxins play a role in the development of the disease. The most important virulence factors include cytolethal distending toxin (CDT) and hemolysin, and the major cell defense mechanisms include superoxide dismutase and membrane factors (11).
Some genes have recently been recognized as responsible for the expression of pathogenicity. In this study, flaA (12), cadF (13) and docC (14) were selected as pathogenic genes responsible for the expression of adherence and colonization, ciaB (15) as a pathogenic gene responsible for the expression of invasion, cdtB as a pathogenic gene responsible for the expression of cytolethal distending toxin production (16), while wlaN (14) and cgtB (17) were selected as pathogenic genes responsible for the expression of Guillain- Barre syndrome (GBS).
2. Objectives
The objective of this study was to analyze the virulence genes of C. jejuni in order to understand the pathogenesis of this microorganism.
3. Methods
3.1. Bacterial Isolates and Growth Conditions
Twenty-five C. jejuni isolates from Rosario, Argentina were tested in this study. These strains were isolated from human feces and were grown on Campylobacter selective agar plates at 42°C for 48 hours under microaerobic conditions.
3.2. Microbiological Tests
The isolates were identified as C. jejuni based on their morphological and biochemical tests, including catalase, oxidase, H2S (TSI), hippurate hydrolysis, resistance to nalidixic acid and sensitivity towards cephalothin.
3.3. Virulence Genes Characterization
A suspension of 200 μL of 24-hour bacterial culture was treated at 100°C for 10 minutes in a hot water bath and cooled in an ice water bath. The suspension resulting from the thermal shock was centrifuged at 10000 g for five minutes. An extract of 5 μL of the supernatant was used as a DNA matrix for the detection of virulence genes. A polymerase chain reaction (PCR) technique was also used to detect the cadF, flaA, cdtB, docC, ciaB, wlaN and cgtB genes. The primers used are recorded in Table 1. The amplification reactions of the genes encoding virulence factors were carried out in a reaction mixture of 25 μL constituting 1× buffer (Fermentans), 3 mM of Cl2Mg, 250 μM of each deoxynucleotide triphosphate (dNTP), 20 pmoles of each primer and 1,25 U of Taq DNA polymerase (Fermentans). polymerase chain reactions were performed in a DNA thermal cycler (IVEMA, Argentina). The amplification program for these virulence markers involved an initial denaturation at 95°C for four minutes and the cyclic phase was repeated 35 times. Each cycle involved a denaturation at 94°C for one minute, a hybridization of the primers at annealing temperatures (Ta) for one minute and an elongation phase at 72°C for one minute; with a final extension at 72°C for five minutes. Details of the Ta are given in Table 1.
| VirulenceFactora | Primers | Oligonucleotide Sequence | Ta, °C | Amplicon Size, bp | References |
|---|---|---|---|---|---|
| cadF | 48 | 400 | (13) | ||
| cadF-F | TTGAAGGTAATTTAGATATG | ||||
| cadF-R | CTAATCCTAAAGTTGAAAC | ||||
| flaA | 52 | 1713 | (12) | ||
| flaA-F | ATGGGATTTCGTATTAACAC | ||||
| flaA-R | CTGTAGTAATCTTAAAACATTTTG | ||||
| cdtB | 55 | 495 | (16) | ||
| cdtB-F | GTTGGCACTTGGAATTTGCAAGGC | ||||
| cdtB-R | GTTAAAATCCCCTGCTATCAACCA | ||||
| ciaB | 50 | 1165 | (13) | ||
| ciaB-F | TTTCCAAATTTAGATGATGC | ||||
| ciaB-R | GTTCTTTAAATTTTTCATAATGC | ||||
| docC | 50 | 1835 | (14) | ||
| docC-F | TGAGCTACGCTATCATTG | ||||
| docC-R | GCTTACGCTATGGGTTGG | ||||
| wlaN | 55 | 330 | (14) | ||
| wlaN-F | TGCTGGGTATACAAAGGTTGTG | ||||
| wlaN-R | ATTTTGGATATGGGTGGGG | ||||
| cgtB | 52 | 561 | (17) | ||
| cgtB-F | TAAGAGCAAGATATGAAGGTG | ||||
| cgtB-R | GCACATAGAGAACGCTACAA |
Abbreviations: bp, bases pair; Ta, annealing temperature.
aGene encoding virulence factor.
Proper negative controls with all components of the PCR mix except for DNA template were included with each PCR run. The revelation of the amplification products was carried out on agarose gel at 1.5% with 0.5μg/ml of ethidium bromide. The estimation of the size of the amplicon was carried out by comparing them with the bands of a molecular weight marker (Thermo Scientific).
4. Results
A total of 30 C. jejuni isolates were screened for the presence of seven virulence-associated genes (data not shown).
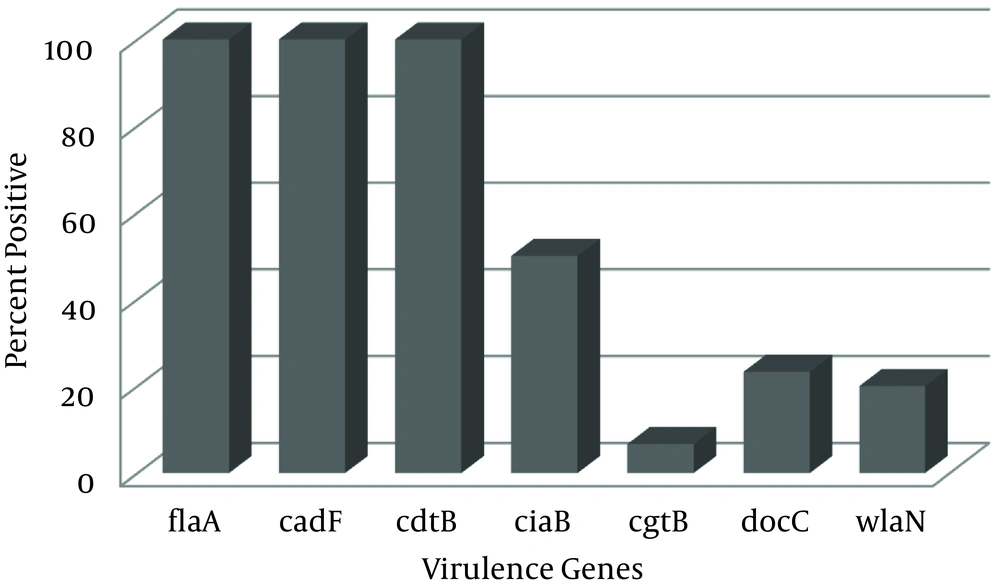
Among these C. jejuni strains, prevalence of cdtB, cadF and flaA virulence genes was 100% (30/30). The frequency of the ciaB gene among C. jejuni strains was 50% (15/30). Furthermore, the presence of docC, wlaN, and cgtB genes was 23.3%, 20% and 6.7%, respectively. Detailed prevalence of the seven virulence genes in C. jejuni recovered from clinical isolates are shown in Figure 1.
Compared with the gene occurrence pattern among the 30 C. jejuni isolates from human feces, nine gene occurrence patterns were detected. It was also shown that all the isolates tested possessed three or more of the seven virulence-associated genes (Table 2).
| Number | Patterns | No. of Isolates |
|---|---|---|
| 1 | flaA, cadF, cdtB, ciaB, docC, wlaN | 1 |
| 2 | flaA, cadF, cdtB, ciaB, docC, cgtB | 1 |
| 3 | flaA, cadF, cdtB, ciaB, wlaN | 3 |
| 4 | flaA, cadF, cdtB, ciaB, docC | 4 |
| 5 | flaA, cadF, cdtB, ciaB | 6 |
| 6 | flaA, cadF, cdtB, wlaN | 2 |
| 7 | flaA, cadF, cdtB, cgtB | 1 |
| 8 | flaA, cadF, cdtB, docC | 1 |
| 9 | flaA, cadF, cdtB | 11 |
| Total | 30 |
5. Discussion
When colonizing the intestines, enteric campylobacters are predicted to express several putative virulence factors. Some genes determine the expression of these virulence factors, which are generally implicated in the processes of adhesion and invasiveness of this pathogen.
As a first step, colonization of the intestine requires the ability to move into the mucus layer covering the intestinal cells. Campylobacter motility is conferred by the polar flagella, which together with their ‘cork-screw’ shape allow them to efficiently penetrate this mucus barrier (18-20). The most important virulence factor that has been studied and well characterized in Campylobacter was the flagellin, which is encoded by the flaA gene (21). The flaA and flaB genes constitute the locus of flagellin; however, molecular genetics research has revealed that flaA is essential for colonization, whereas the flaB gene is not (22).
As mentioned before, it is known that flaA, cadF and ciaB genes, studied in this work are involved in adhesion and invasiveness of Campylobacter spp. (23, 24). Moreover, it has been seen that ciaB gene has been associated with cell invasion, as it encodes for the secretion of a protein necessary for the invasion of epithelial cells (8). The cadF gene, in turn, encodes a protein that interacts with the host’s fibronectin matrix, which is necessary for colonization of the cell surface (25).
In this study, a number of putative virulence and toxin genes were studied, including flaA, cadF and ciaB genes, that are involved in adhesion and colonization of the host’s gut (13, 14). All tested C. jejuni isolates were positive for the flaA gene. Previous studies showed that the detection rate of flaA gene was 95% (26) and 100% (27, 28).
A study conducted by Rozynek et al. (29) on C. jejuni coli isolates showed that all analyzed strains possessed the cadF gene. In a similar study, all isolates with human origin had the cadF gene (30). Our results detected the cadF gene in 100% of the tested strains, as was observed by Biswas et al., Datta et al. and Gripp et al. (27, 31, 32). The prevalence of flaA and cadF gene in all the isolates indicates a pathogenic potential since both genes play an important role in Campylobacter pathogenesis.
On the other hand, the detection rate of ciaB was 50%. In studies conducted by Biswas et al., Datta et al. and Hyun-Ho Cho et al., the ciaB gene was detected in a 92.31%, 98.2% and 87.5%, respectively (27, 28, 31).
This study was performed to investigate the CDT-encoding gene (cdtB) of C. jejuni from human clinical samples. The cdt gene cluster consists of three adjacent genes (cdtA, cdtB, and cdtC). The CDT toxin is composed of CdtB protein, as the enzymatically-active subunit and two hetero-dimeric subunits (CdtA and CdtC), which are responsible for the holotoxin binding to the cell membrane (33).
The cdtB was detected in 100% of the isolates tested in this study, which is consistent with previous reports by Asakura et al., Thakur et al. and Gripp et al. (26, 32, 33).
Two Guillain- Barre syndromes-associated genes (cgtB and wlaN) were detected by the PCR method, showing that cgtB gene was detected in 6.7% and wlaN gene was detected in 20.0% of the isolates. The cgtB and wlaN gene product as β-1, 3-galactotransferase is responsible for the specific LOS structure. LOS, similar to gangliosides in neurons, is thought to be a critical factor in the triggering of GBS and Miller-Fisher syndrome neuropathies after C. jejuni infection (3, 17). The higher prevalence of these genes might be associated with GBS in humans.
The rate of the presence of wlaN gene is similar to that obtained in other studies (27). However, Hyun-Ho Cho et al. showed 71.7% prevalence for the cgtB gene among isolates from human sources (28).
Finally, in some enteric pathogens, such as C. jejuni, chemotaxis is important for pathogenesis and colonization of the host. Chemotaxis allows motile bacteria to navigate depending on the extracellular chemical composition. Bacteria are either attracted or repelled by chemicals sensed by trans-membrane Methyl-Accepting Chemotaxis proteins (MCP). Among the identified determinants, there was a gene encoding a MCP (docC), presumably required for proper chemotaxis to a specific environmental component (34). In our work, docC gene was present in 23.3% of the strains. Little is known about the prevalence rate of this gene. Hyun-Ho Cho et al. reported the prevalence rate of this gene in C. jejuni isolates from humans to be 19.6% (28).
In conclusion, several virulence factors have been documented for Campylobacter spp., which could contribute to its motility, intestinal colonization and invasion.
The present study showed the high prevalence of the cadF, flaA and cgtB genes among C. jejuni isolated from human feces. Furthermore, the isolates showed a pattern, which is different from other studied pathogenic genes. Moreover, this work enhances knowledge on the prevalence of virulence factors in C. jejuni isolated from human feces, and the contribution of these characteristics to clinico-epidemiological monitoring.