1. Background
Streptococcus pneumoniae is a significant cause of mortality and morbidity worldwide; the estimated global mortality rate for S. pneumoniae in 2015 was reported as 45 deaths (29 - 56) per 100000 children younger than five years (1). Pneumococcal nasopharyngeal colonization, reported in 27- 65% of children, is a potential cause of severe pneumococcal infections (e.g., pneumonia, meningitis, and sepsis) and a source of transmission of the pathogen (2).
In 2000, the first pneumococcal conjugate vaccine (PCV7), comprising the serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F, was licensed by the American Food and Drug Administration (FDA). Since then, PCV10 and PCV13 have also been introduced, each covering additional serotypes to those covered by PCV7 (3). Following the introduction of pneumococcal vaccines, a significant reduction in incidence, hospitalization, and mortality rates related to pneumococcal diseases was observed in high-income and non-high-income countries (4). However, the available vaccines only include the serotypes responsible for severe pneumococcal disease in developed countries, and the serotype-specific immunity induced by vaccination has led to a relative increase in non-vaccine serotypes in other geographic regions (5, 6). Local and regional detection of invasive pneumococcal serotypes and monitoring the changes over time is essential to investigate the most common invasive serotypes and their coverage by current pneumococcal vaccines.
2. Objectives
Considering the novelty of study on pneumococcal serotypes in Iran and the necessity of its performance, this study aimed to detect S. pneumoniae serotypes in children with pneumonia, sepsis, and meningitis by PCR method and estimate their coverage with different pneumococcal vaccines.
3. Methods
A cross-sectional study was conducted on 563 blood, and cerebrospinal fluid (CSF) specimens collected from children aged one month -18 years and suspected sepsis, pneumonia, and meningitis hospitalized at Mofid Children's Hospital and other regional hospitals from March 20, 2012 to March 10, 2013. Patients with confirmed immunodeficiency, diagnosed underlying disease, nosocomial infection, and children with chronic cough (more than two weeks) were excluded.
Clinical specimens, including 1.5 - 2 ml blood and 1 ml CSF, were obtained by a sterile procedure according to the recommendations for collecting specimens for laboratory diagnosis of S. pneumoniae and Haemophilus influenza (7). Blood specimens were injected in culture broth and mixed gently, and CSF specimens were collected in a sterile screw-capped tube. The samples were transported to the laboratory within 4 - 6 hrs. The culture was done in BACTEC 9120 BD system. Confirmatory tests identified blood and CSF positive culture specimens. DNA extraction was performed by Roche (High pure PCR template preparation kit, Cat No: 11796828001) kit, and S. pneumoniae was confirmed by a specific primer of the cps gene and PCR technique. Confirmed serotyping was done on confirmed Pneumococcus specimens by Multiplex PCR. Specific cps gene primers and PCR solution for each Multiplex PCR group are shown in Tables 1 and 2.
| No | Primer Name | Sequence Primer | Size Primer (bp) |
|---|---|---|---|
| 1 | CpsA F | GCAGTACAGCAGTTTGTTGGACTGACC | 27 |
| 2 | CpsA R | GAATATTTTCATTATCAGTCCCAGTC | 26 |
| 3 | 1 F | CTCTATAGAATGGAGTATATAAACTATGGTTA | 32 |
| 4 | 1 R | CCAAAGAAAATACTAACATTATCACAATATTGGC | 34 |
| 5 | 4 F | CTGTTACTTGTTCTGGACTCTCGATAATTGG | 31 |
| 6 | 4R | GCCCACTCCTGTTAAAATCCTACCCGCATTG | 31 |
| 7 | 5 F | ATACCTACACAACTTCTGATTATGCCTTTGTG | 32 |
| 8 | 5 R | GCTCGATAAACATAATCAATATTTGAAAAAGTATG | 35 |
| 9 | 6A/6B/6C F | AATTTGTATTTTATTCATGCCTATATCTGG | 30 |
| 10 | 6A/6B/6C R | TTAGCCGAGATAATTTAAAATGATGACTA | 29 |
| 11 | 7f/7a F | CCTACGGGAGGATATAAAATTATTTTTGAG | 30 |
| 12 | 7f/7a R | CAAATACACCACTATAGGCTGTTGAGACTAC | 30 |
| 13 | 7C F | CTATCTCAGTCATCTATTGTTAAAGTTTACGACGGG | 36 |
| 14 | 7C R | GAACATAGATGTTGAGACATCTTTTGTAATTC | 32 |
| 15 | 8 F | GATGCCATGAATCAAGCAGTGGCTATAAATC | 31 |
| 16 | 8 R | ATCCTCGTGTATAATTTCAGGTATGCCACC | 30 |
| 17 | 9V F | CTTCGTTAGTTAAAATTCTAAATTTTTCTAAG | 32 |
| 18 | 9V R | GTCCAATACCAGTCCTTGCAACACAAG | 27 |
| 19 | 10A F | GGTGTAGATTTACCATTAGTGTCGGCAGAC | 30 |
| 20 | 10A R | GAATTTCTTCTTTAAGATTCGGATATTTCTC | 31 |
| 21 | 11A F | GGACATGTTCAGGTGATTTCCCAATATAGTG | 31 |
| 22 | 11A R | GATTATGAGTGTAATTTATTCCAACTTCTCCC | 32 |
| 23 | 12f F | GCAACAAACGGCGTGAAAGTAGTTG | 25 |
| 24 | 12f R | CAAGATGAATATCACTACCAATAACAAAAC | 30 |
| 25 | 14 F | GAAATGTTACTTGGCGCAGGTGTCAGAATT | 30 |
| 26 | 14 R | GCCAATACTTCTTAGTCTCTCAGATGAAT | 29 |
| 27 | 15A F | ATTAGTACAGCTGGAATATCTCTTC | 25 |
| 28 | 15A R | GATCTAGTGAACGTACTATTCCAAAC | 26 |
| 29 | 15B/C F | TTGGAATTTTTTAATTAGTGGCTTACCTA | 29 |
| 30 | 15B/C R | CATCCGCTTATTAATTGAAGTAATCTGAACC | 31 |
| 31 | 16 F | CTGTTCAGATAGGCCATTTACAGCTTTAAATC | 32 |
| 32 | 16 R | CATTCCTTTTGTTATTAGTGCTAGTTCATCC | 31 |
| 33 | 17 F | TTCGTGATGATAATTCCAATGATCAAACAAGAG | 33 |
| 34 | 17 R | GATGTAACAAATTTGTAGCGACTAAGGTCTGC | 32 |
| 35 | 18c F | CTTAATAGCTCTCATTATTCTTTTTTTAAGCC | 32 |
| 36 | 18c R | TTATCTGTAAACCATATCAGCATCTGAAAC | 30 |
| 37 | 19 F F | GTTAAGATTGCTGATCGATTAATTGATATCC | 31 |
| 38 | 19 F R | GTAATATGTCTTTAGGGCGTTTATGGCGATAG | 32 |
| 39 | 19A F | GAGAGATTCATAATCTTGCACTTAGCCA | 28 |
| 40 | 19A R | CATAATAGGTACAAATGACTCATCGCC | 27 |
| 41 | 20 F | GAGCAAGAGTTTTTCACCTGACAGCGAGAAG | 31 |
| 42 | 20 R | CTAAATTCCTGTAATTTAGCTAAAACTCTTATC | 33 |
| 43 | 22f F | GAGTATAGCCAGATTATGGCAGTTTTATTGTC | 32 |
| 44 | 22f R | CTCCAGCACTTGCGCTGGAAACAACAGACAAC | 32 |
| 45 | 23f F | GTAACAGTTGCTGTAGAGGGAATTGGCTTTTC | 32 |
| 46 | 23f R | CACAACACCTAACACACGATGGCTATATGATTC | 33 |
| 47 | 31 F | GGAAGTTTTCAAGGATATGATAGTGGTGGTGC | 32 |
| 48 | 31 R | CCGAATAATATATTCAATATATTCCTACTC | 30 |
| 49 | 33f F | GAAGGCAATCAATGTGATTGTGTCGCG | 27 |
| 50 | 33f R | CTTCAAAATGAAGATTATAGTACCCTTCTAC | 31 |
| 51 | 34 F | GCTTTTGTAAGAGGAGATTATTTTCACCCAAC | 32 |
| 52 | 34 R | CAATCCGACTAAGTCTTCAGTAAAAAACTTTAC | 33 |
| Primers | Addition Primers (µL) | DNA Sample (µL) | Distilled Water (µL) | The Final Volume (µL) |
|---|---|---|---|---|
| 19A | 2 | 3 | 7 | 20 |
| 19F | 2 | |||
| 6A/B | 2 | |||
| 1 | 2 | |||
| CPS | 2 | |||
| 5 | 2 | 3 | 9 | 20 |
| 14 | 2 | |||
| 7F/A CPS | 2 | |||
| 5 | 2 | |||
| 23F | 3 | 3 | 4 | 20 |
| 7F | 4 | |||
| 11A | 2 | |||
| 33F | 2 | |||
| cps | 2 | |||
| 16F | 4 | 3 | 6.5 | 20 |
| Sg18 | 2.5 | |||
| 35B | 2 | |||
| CPS | 2 | |||
| 8 | 3 | 3 | 4 | 20 |
| 15B/C | 3 | |||
| 38 | 3 | |||
| 31 | 4 | |||
| 1 | 3 | 3 | 5 | 20 |
| 10A | 3 | |||
| 35F | 3 | |||
| 34 | 3 | |||
| 20 | 2 | 3 | 7 | 20 |
| 7C | 2 | |||
| 17F | 2 | |||
| 15B/C | 2 | |||
| cps | 2 | |||
| 4 | 2 | 3 | 9 | 20 |
| 14 | 2 | |||
| 12 | 2 | |||
| 9V | 2 | |||
| CPS | 2 | 2 | 16 | 20 |
| 15A | 2 | 2 | 16 | 20 |
| 22F | 2 | 2 | 16 | 20 |
Data regarding patients' characteristics and PCR results were recorded in separate checklists and analyzed using SPSS ver 22. Results were reported as numbers (percent).
4. Results and Discussion
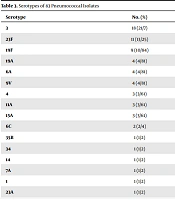
The PCR results indicated that 83 out of 563 (14.7%) specimens were positive for S. pneumoniae. Multiplex PCR typed 67 of 83 (80.7%) isolated pneumococci, however 16 samples (19.3%) were non-typeable (Table 3). Overall, 16 serotypes were responsible for the confirmed S. pneumoniae infections, the most prevalent serotypes being serotype 3 (21.7%), 23F (13.25%), and 19F (10.84%).
| Serotype | No. (%) |
|---|---|
| 3 | 18 (21/7) |
| 23F | 11 (13/25) |
| 19F | 9 (10/84) |
| 19A | 4 (4/81) |
| 6A | 4 (4/81) |
| 9V | 4 (4/81) |
| 4 | 3 (3/61) |
| 11A | 3 (3/61) |
| 15A | 3 (3/61) |
| 6C | 2 (2/4) |
| 35B | 1 (1/2) |
| 34 | 1 (1/2) |
| 14 | 1 (1/2) |
| 7A | 1 (1/2) |
| 1 | 1 (1/2) |
| 23A | 1 (1/2) |
| Nontyeable | 16 (19.3) |
| Total | 83 (100) |
Compared with other studies performed in Iran, our findings showed some differences regarding prevalent pneumococcal serotypes. According to a study in Northeast Iran, serotypes 23F, 19F, 19A, 1, 14, and serogroup 6A/B were the most common types among children with suspected meningitis (8). Serotypes 23F, 14, 3, 19, and 19A were reported as the most prevalent in children with invasive and non-invasive pneumococcal infections admitted to Shariati Hospital, Tehran (9). Moreover, in a study on clinical samples obtained from children with an invasive pneumococcal disease from several hospitals in Tehran, 23F, 19F, 19A, and 9V as the most common serotypes (10).
Studies from other countries have reported a wide variety of prevalent serotypes. In a study conducted in 60 hospitals from 11 Asian countries, the most common typeable serotypes were 19F (23.5%), 23F (10%), and 19A (8.2%); and non-typeable serotypes accounted for (5.4%) of isolates (11). On the other hand, a Turkish study performed on 167 children with invasive pneumococcal disease reported 19F, 1, 3, and 19A as the most common invasive pneumococcal serotypes (12). The differences in pneumococcal serotype distribution can be attributed to several factors such as geographic regions, type of pneumococcal disease (invasive or non-invasive), study population characteristics, source of clinical specimens, and PCR method.
Iranian national immunization program does not include routine pneumococcal vaccination. Accordingly, there is lower PCV7, PCV10, and PCV13 coverage than some other countries (11-13). Interestingly, pneumococcal vaccine coverage reported by Abdoli et al and Houri et al were higher than our findings (8, 10). Differences in pneumococcal serotypes distribution might be explained by geographical and temporal changes and PCR techniques. Based on our results, PCV-13 is the most influential vaccine against common invasive pneumococcal serotypes, covering 66.2% of serotypes detected by PCR. As most samples were collected from patients hospitalized in Mofid children’s hospital, one of the primary referral pediatric centers in Iran, PCV-13 might be considered for inclusion in the national immunization program. However, further studies in different parts of Iran are recommended, as serotype distribution may change over time.
The study's limitation was difficulties in at time specimen collection before the start of antibiotics We organized our educated physicians to obtain standard specimens for serotyping at the time of admission. Our study's advantages were using clinical isolates that represent invasive pneumococcal serotypes in our society.
5. Conclusions
We found that serotypes 3, 23F, and 19F accounted for almost 46% of invasive pneumococcal isolates. As per relatively high representation of prevalent serotypes in PCV-13, this vaccine is the preferred choice in Tehran, Iran. More research on serotypes in different geographical regions allows the design of a nationwide vaccination program.
The Ethics Committees of Shahid Beheshti University of Medical Sciences and Tehran University of Medical Sciences, Tehran, Iran approved the study (Ethics code: 1391-1-87-9954-10064).