1. Background
Bloodstream infection is a leading cause of morbidity and mortality in neonatal intensive care units. The microbiological diagnosis of sepsis in newborns (NBs) is a difficult task, because blood culture (BC), which is currently the gold standard for diagnosing NBs (1) has the disadvantage of producing an elevated percentage of false negatives (2). Previous studies have shown that 47.1% of NBs who begin antimicrobial treatment have negative culture results, and some analysis concluded that sepsis is diagnosed excessively (3). In addition, diagnoses based on clinical criteria and experience favor overdiagnosis and antibiotic abuse (4, 5).
Many evaluations of the available diagnostic tests have attempted to solve the problem described above, but their success has been limited (6). Unfortunately, the decision to delay treatment until obvious signs and symptoms of sepsis occur, leads to the risk of preventable mortality (7).
However, few studies have addressed the clinical impact of the use of LC-SF (a technique of molecular biology for rapid detection of bacterial and fungal infections) in NBs.
2. Objectives
In the present study, we aimed to evaluate the accuracy of the LC-SF assay for detection of bacterial and fungal agents compared to BC and CRP, using LC–SF like a routine clinical practice in five hospitals. We evaluated the impact of used LC-SF on some outcome variables of NBs with NS.
3. Materials and Methods
First, the study was performed prospectively in patients admitted to the Central South Hospital of Petroleos Mexicanos, the Gynecological-Obstetrics Hospital number 4 of the Mexican Institute of Social Security; the Dalinde Hospital (Private institution); and the Monterrey Nuevo Leon University Hospital, and National Institute of Perinatology.
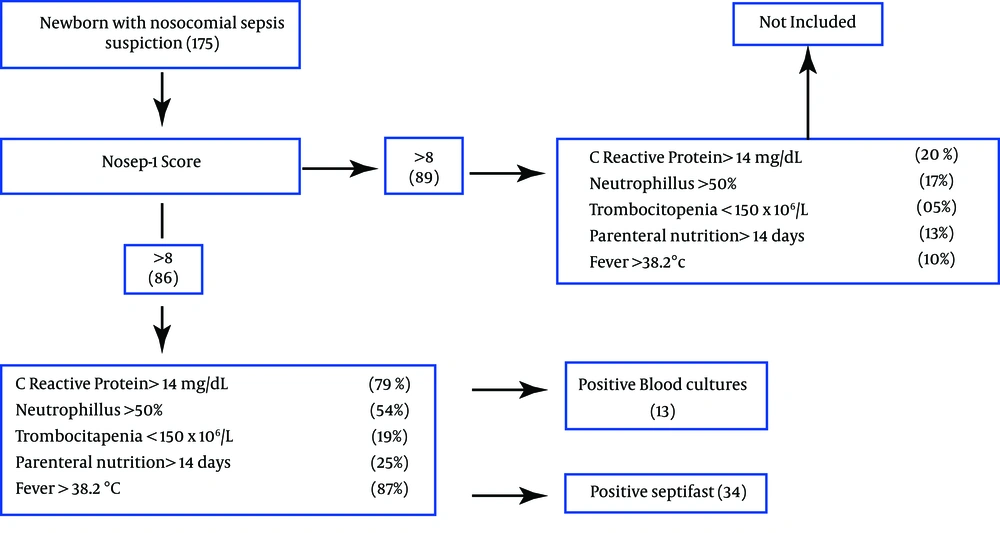
All patients with clinical sepsis suspiction and those presenting a score of > 8 points on the NOSEP-1 scale were included in the study. NOSEP-1 scale is composed by C-reactive protein > 14 mg/dL (5 points), neutrophils >50% (3 points), thrombocytopenia < 150 × 106/L (5 points), parenteral nutrition > 14 days (6 points), fever > 38.2°C (5 points) (8); It was previously validated for our population (9). After the NOSEP-1 punctuation criterion was satisfied, two blood samples were collected from each patient prior to antimicrobial treatment. The blood samples underwent culturing and LC-SF assay in parallel. Only one episode per patient was considered; we did not exclude NBs who had previously used prophylactic or antimicrobial treatment (Figure 1).
For the LC-SF test, DNA was extracted (using sterile technique) from a 1-mL blood sample in a tube containing EDTA as an anticoagulant. The sample was mechanically lysed using the MagNA Lyser Instrument (Roche Diagnostics GmbH, Germany). The SeptiFast Prep® MGRADE Kit was used to purify the DNA, and the samples were incubated with a protease and a lysis buffer at 56 ºC to liberate the DNA and protect it from DNases. Subsequently, the binding buffer was added, and the solution was transferred to a fibreglass column in which the human, bacterial and fungal genomic DNA was retained while the remainder of the material was removed by two washes. Finally, the nucleic acids were eluted in 300 μL of the solution and used as templates for the amplification reaction.
The LightCycler® SeptiFast Test MGRADE Kit was used for amplification reactions containing Taq polymerase and primers/specific probes designed to detect Gram-positive bacteria, Gram-negative bacteria and yeasts. The purified DNA samples and the other reaction components were added to the capillaries (100 μL) of the LightCycler® MGRADE, and the PCR amplification was performed with the LightCycler® 2.0 equipment (Roche Diagnostics GmbH, Germany). A melting curve was then generated for the controls and the amplified samples and was analysed automatically by specific identification software (SeptiFast Identification Software, Roche Diagnostics GmbH, Germany). We used both positive and negative controls by assay.
The blood cultures were performed using sterile technique. Each 1-mL sample was inoculated in a BacT/ALERT bottle (BioMerieux SA) and incubated using an automated system made by the same company. The blood cultures were requested as part of the protocol to study children with suspected sepsis.
Additionally, the impact of LC-SF results on hospitalization days, mortality, mechanical ventilation days and days of antibiotics use were analyzed retrospectively. We compared 86 NBs with result of LC-SF vs 86 NBs collected from clinical records with nosocomial sepsis suspiction with BC results. The evaluation of costs was not considered to study.
3.1. Statistical Analysis
The demographic characteristics of the patients were analyzed using descriptive statistics, mainly averages and medians. The efficacy of SeptiFast for microbial identification was analyzed by calculating its sensitivity, specificity and predictive values using the blood culture like gold standard. The concordance between the two types of microbial identification tests, blood culture and LC-SF was analyzed by calculating the Kappa coefficient.
3.2. Ethics
The study was approved by the institutional research and ethics committees.
4. Results
A total of 86 samples from 86 NBs with suspiction of nosocomial sepsis were included in the study. Fifty nine (68.6%) patients were male and 27 (31.3%) female. Among the patients, 43.7% weighed >1500 g, and 56.3% weighed <1500 g. A gestational age of more than 35 weeks was reached by 43.7% of the NBs, whereas 56.3% were born at or before 35th week.
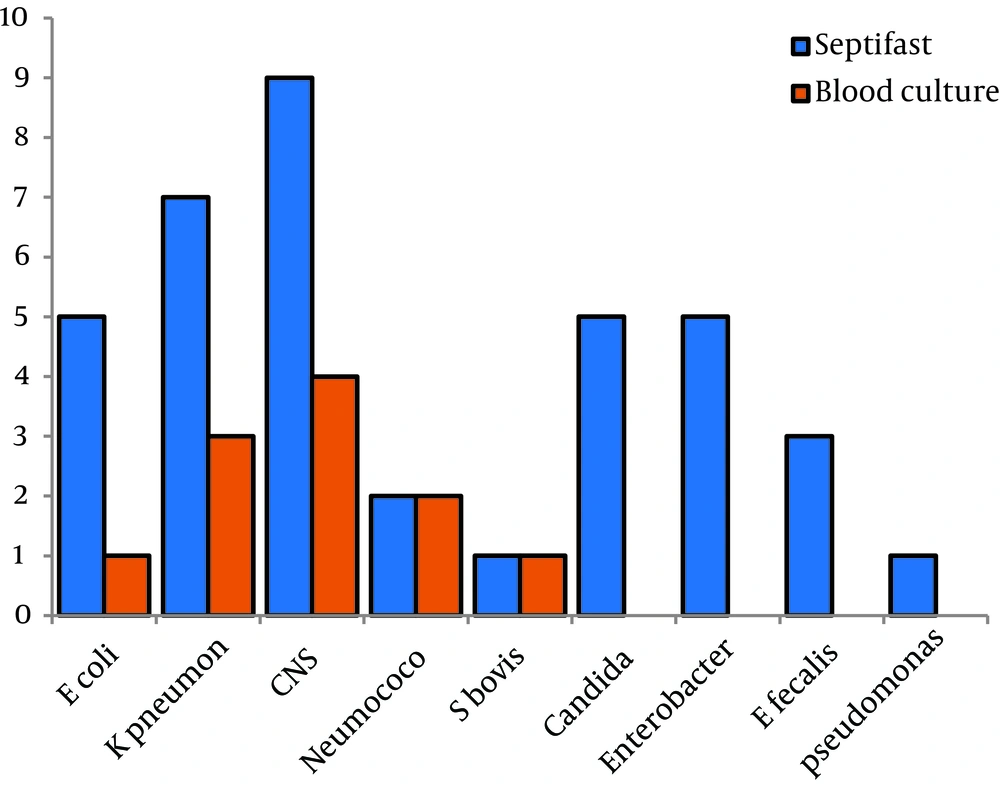
Results of 86 episodes (paired blood samples tested by blood culture and SeptiFast) were evaluated. 13/86 (15.1%) blood cultures were positive, and 34/86 (39.5%) were positive with SeptiFast tests (Table 1). The sensitivity of SeptiFast test compared with blood culture was 0.69, specificity 0.65, positive predictive value 0.69 and the negative predictive value 0.65. Results for isolates from all episodes are shown in Figure 2, compared with blood culture, SeptiFast showed similar or higher percentages of isolates for all the studied microorganisms, with differences: E. coli (20%), K. pneumoniae (42.8%), SCN (44.4%), Pneumococcus (0%), S. bovis (0%), Candida spp, Enterobacter spp, E. faecalis and Pseudomonas spp (100%). Polymicrobial infection rates were found in 4.6%. with SeptiFast and 0% with blood culture (Table 2).
Kappa index concordance between LC-SF and BC was 0.21; while Kappa index concordance between LC-SF and CRP was 0.17 (Table 3). The response time was 6-8 hours using the LC-SF test and from 4 to 7 days using blood culture detection. Hospitalization days, days of antimicrobial use, and number of subjects died diminished meaningful (Table 4).
| Patient (n = 38) | Septifast Results | Blood Culture Results |
|---|---|---|
| Negative | ||
| Negative | ||
| Negative | ||
| Negative | ||
| Negative | ||
| Negative | ||
| Negative | ||
| Negative | ||
| Negative | ||
| Negative | ||
| Negative | ||
| Negative | ||
| Negative | ||
| Negative | ||
| Negative |
a Negative results in both, septifast and blood cultures are not considered in the table (n = 48). The sensitivity of SeptiFast test compared with blood culture was 0.69, specificity of 0.65, positive predictive value was 0.69 and the negative predictive value was 0.65.
| Test | Positive BC (n = 13) | Negative BC (n = 73) |
|---|---|---|
| SCN (5) | ||
| Escherichia coli (4) | ||
| Staphylococcus epidermidis (2) | ||
| Total = 9 | ||
| SCN + | ||
| SCN + Enterococcus faecalis | ||
| Total = 25 | ||
| 48 negative | ||
| Total = 4 |
a Using blood culture, 8 Gram-positive and 5 Gram-negative bacteria were isolated, whereas DNA sequences from 15 Gram-positive bacteria, 18 Gram-negative bacteria and 5 yeasts were identified using LC-SF. In two cases, LC-SF identified Streptococcus pneumoniae, which is a rare infectious agent in our neonatal population. In both cases, the blood culture results confirmed the presence of S. pneumoniae five days after its detection with molecular biology.
| BC (+) | BC (-) | OA | EA | KI | ES | 95% CI | CF | |
|---|---|---|---|---|---|---|---|---|
| 9 | 25 | 0.66 | 0.57 | 0.21 | 0.11 | -0.02,0.44 | Weak | |
| 4 | 48 | |||||||
| 13 | 73 |
a Abbreviations: BC, blood culture; CF, concordance factor; EA, expected average; KI, Kappa index;. OA, observed average.
| Variable | Septifast | Blood culture | P Value |
|---|---|---|---|
| 1.098 + 250 | 1.060 + 270 | 0.6 | |
| 30.6+ 3 | 29.6 + 2.1 | 0.07 | |
| 48+22 | 65 + 40 | 0.05 | |
| 15+10 | 23 + 9.2 | 0.05 | |
| 11 +6 | 13+ 7 | 0.9 | |
| 7 (8.1) | 12 (13.9) | 0.05 |
5. Discussion
The understanding of neonatal sepsis as a disease with a critical public health impact is based on the identification of factors that magnify the problem by favoring long hospital stays and increasing the use of diagnostic and treatment resources (10). The high degree of variation in clinical judgement adds to the difficulty of sepsis microbial identification and is a key reason behind overdiagnosis and the indiscriminate use of antimicrobials (11). As the golden standard validation technique, BC has poor sensitivity, favors confusion and contributes to the lack of diagnostic specificity that is characteristic of the systemic inflammatory response syndrome (SIRS) in NBs. The disadvantages of relying on BC to diagnose sepsis are exemplified by the adoption of terminology to justify antibiotic treatment, such as the concept of “sepsis without an isolated germ” applied to a child who fulfills the criteria for SIRS but lacks an isolated microorganism. The main consequence of this variability is the abuse of antibiotics in an effort to prevent mortality due to sepsis (12).
LC-SF is a multiplex real-time PCR assay based on specific probes that detect and identify DNA sequences from 25 microorganisms (primarily Gram-positive bacteria, Gram-negative bacteria and yeasts) that represent 90% of the species isolated from septic patients. Detection of virus or Mycobacterias is not possible with LC-SF; and the microbial sensitivity to antibiotics and antibiogram is not available. This technique has led to improved rates of detection of the causative bacteria for diseases such as infectious endocarditis, sepsis in neutropenic patients with hematological malignancies, bacteremias and fungemias in specific groups (13-16).
There have been relatively few reports of LC-SF used to diagnose NBs. The reported sensitivity levels of 79-90.5%, specificity of 87%, negative predictive value of 83% and positive predictive value of 83% compared with blood culture (17-19) are higher than the values from the present study. A meta-analysis by Chang et al. concluded that the SeptiFast assay had a sensitivity of 75% and a specificity of 92% for detecting bacteremia or fungemia (20). Nonetheless, the identification capacity of SeptiFast was greater than that of blood culture both in the overall data analysis and in the analyses of individual microorganisms, especially yeasts and Pseudomonas.
The early detection of microorganisms using a technique based on the identification of nucleic acids will promote the prompt and appropriate use of antimicrobials, which decreases mortality. The prompt detection of the causative agent is crucial for the proper treatment of an infection.
Some limitations of the present study should be noted. First, we did not examine the possibility of contamination during the identification process, a factor that impacts the efficacy of most assays according to previous studies (21), because the conditions for inclusion in our study ensured that none of the isolates were considered contaminated. Coagulase-negative Staphylococci were considered possible agents of sepsis in these children because these bacteria are frequently isolated in Mexican population (22). Additional support for the absence of contamination in the present work is found in another study that evaluated LC-SF and reported no cases of contamination; those researchers applied the standardised criteria for SIRS developed by a consensus of the American College of Chest Physicians/Society of Critical Care Medicine (ACCP/SCCM), which ensures compliance with practices that minimise contamination (17). Previously an algorithm for interpretation of BC and LC-SF results was reported by Lucignano B et al. there were criteries to consider a pathogen and a contaminant; these criteries were the same as in our research (23). Second, some authors, such as Lodes (17) have reported that the SeptiFast test detects polymicrobial infection at rates as high as 11.4%, a higher (4.6%) rate than that was found in our study. This discrepancy might be explained by the Lodes study’s examination of surgical patients from an intensive care unit, where the polymicrobial infection rate is high.
Unlike other studies, our findings influenced the treatments administered to our subjects, which substantiates our results. As a prime example, the identification of positive fungal LC-SF results led to specific patient treatments, which resulted in clinical improvement. Therefore, analyzing the follow-up care of our subjects enabled a more complete evaluation of the usefulness of the LC-SF test in terms of the length of hospitalisation, use of antimicrobials and duration of recovery.
Microbial identification in neonates is a relatively understudied topic that has generated some controversy because of possible incorrect decision making due to false positives. Therefore, the present work provides valuable support for the use of multiplex PCR as an identification method that can be performed together with BC, has the advantage of a 6 hour response time and allows the use of more targeted antimicrobial therapy. Similar to scenarios involving patients with negative blood culture results, when patients have negative LC-SF results but solid clinical data and a high suspicion of SIRS, the clinical criteria will continue to be the mainstay for diagnosis and treatment. The initiation of antimicrobial treatment based on suspicion will most likely not decrease. We predict that the only difference associated with LC-SF-based diagnosis will be a higher percentage of positive test results (compared with BC) and a more rapid response time leading to decision-making based on more solid evidence, which is the ultimate objective of auxiliary discriminatory tools. It is highly unlikely that SeptiFast (or another biomarker) will replace clinical criteria for diagnosis.