1. Background
Russell-Silver syndrome is a rare genetic syndrome categorized as one the primordial dwarfism (PD) syndromes (1, 2). This syndrome was first described because of its growth disturbances and syndromic clinical appearance. The most common clinical features are characteristic triangular face with prominent forehead, postnatal growth failure, body asymmetry, and feeding difficulties (3, 4). Primary clinical guidelines for the diagnosis of Russell-Silver syndrome have been made based on such clinical features. The prevalence of this syndrome has been reported to vary from 1:30000 to 1:100000 children according to these clinical guidelines (5). Recently, according to the development of molecular techniques, the molecular nature of Russell-Silver syndrome has become more evident and new diagnostic algorithms have been established. Evaluating the loss of methylation on 11p15 and maternal uniparental disomy for chromosome 7 (upd(7)mat) are the most common molecular changes that can be assessed in such patients (6, 7). However, the molecular change is determined only in two-thirds of the patients and other cases with typical features of Russell-Silver syndrome remain unknown (8). The loss of methylation on 11p15 has been reported to account for 30% to 60% of the patients (8). While this molecular change has been considered the most common molecular change in children with clinical features of Russell-Silver syndrome.
2. Objectives
We conducted a study to evaluate this molecular alteration in patients who were considered to have this syndrome according to the clinical guidelines.
3. Methods
This case series was approved by Mashhad University of Medical Sciences Committee and conducted at Imam Reza Hospital, Mashhad, Iran. Every child who presented with a clinical diagnosis of Russell-Silver syndrome aged younger than 20 and older than two years was referred to the pediatric endocrinology department for further evaluations. Among children who were referred, only 15 patients were confirmed to have Russell-Silver syndrome clinically according to the Netchine-Harbison clinical scoring system (9). According to these criteria, the clinical diagnosis was made if a child scored at least four out of six clinical criteria including small for gestational age, postnatal growth failure, relative macrocephaly at birth, body asymmetry, protruding forehead and feeding difficulties and/or low body mass index. Other possible differential diagnoses were excluded according to the children features, as the same as those who were not willing to undergo a molecular evaluation or disagreed with filling an informed consent form. A total number of 15 children agreed to enroll in the present study and undergo a molecular evaluation. The children were referred to the Iranian Academic Center for Education, Culture, and Research genetic laboratory. Five mL of peripheral venous blood was taken and DNA was extracted by using the salting-out method. The SALSA® MS-MLPA® BWS/RSS ME030-C3 probemix kit (MRC-Holland) was used for the evaluation of the 11p15 region, H19, IGF2, CDKN1C, and KCNQ1. According to the manufacturer notice, this kit had the capacity for determining both copy number and methylation status of the 11p15 region. Then, capillary electrophoresis was performed and the confirmation of Russell-Silver syndrome was reported to the physician.
4. Results
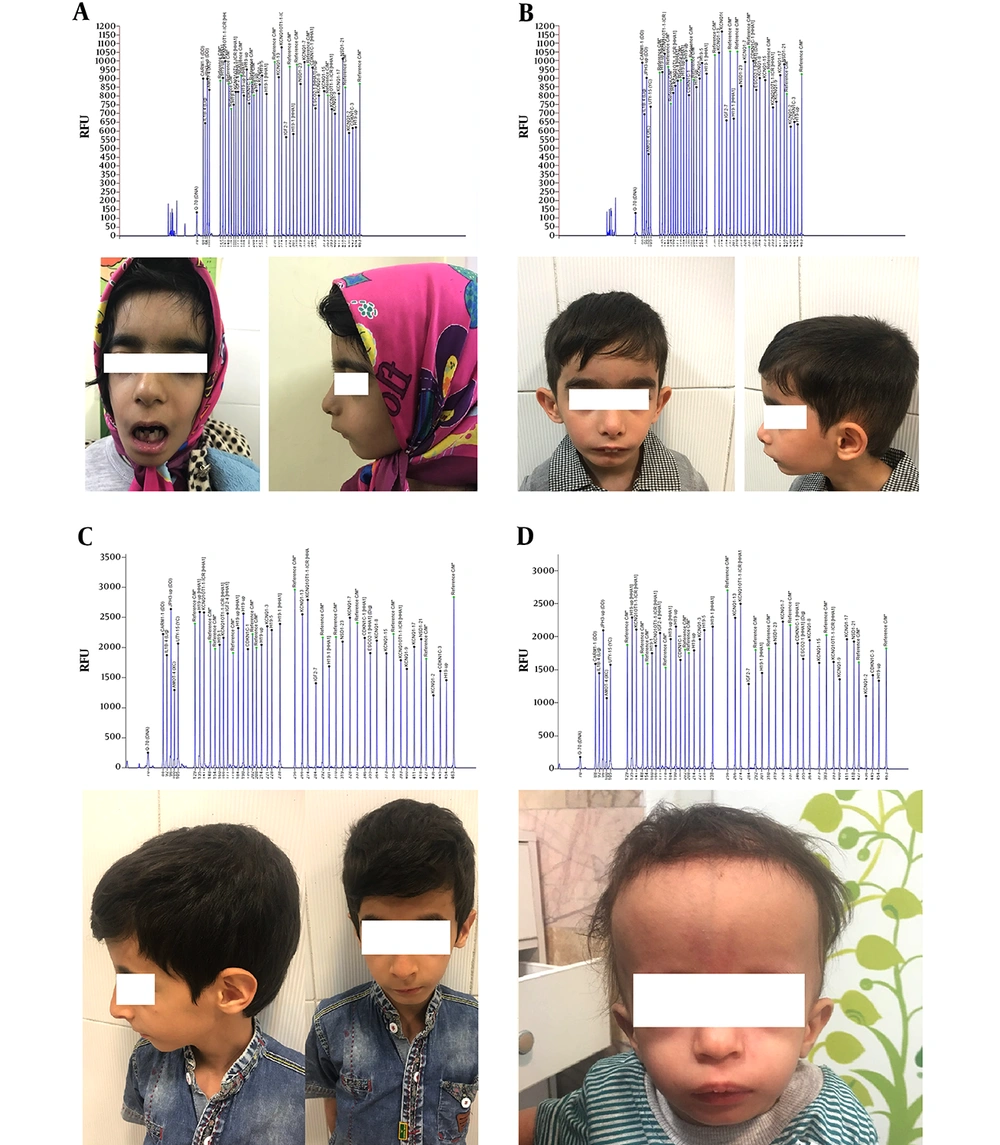
The mean age (standard deviation) of children who were eligible to enroll in this study was 4.5 (1.6) years. Among the 15 children, seven (46.7%) were girls and eight (53.3%) were boys. The mean (standard deviation) values of birth weight and height were 1615 (434) grams and 41.2 (2.7) centimeters, respectively. Only one girl and three boys (26.6%) were considered to have the confirmed molecular diagnosis of Russell-Silver syndrome. All of these children had low set ears, high pitch voice, micrognathia, low body mass index, triangular face, and growth retardation. Among these four children (26.6%), only one had body asymmetry. Two children had findings closely similar to the findings of confirmed cases but their MS-MLPA results came back negative. Overall symptoms in all children are summarized in Table 1 and the pictures and Capillary electrophoresis of four children who had the confirmed molecular diagnosis are shown in Figure 1.
| Patient Number | Age, y | Micrognathia | Triangular Face | Feeding Difficulty | Body Asymmetry | Prominent Forehead | Clinodactyly | High Pitch Voice | Low Set Ear |
|---|---|---|---|---|---|---|---|---|---|
| 1a | 7 | + | + | + | - | + | + | - | + |
| 2a | 3 | + | + | + | - | + | - | + | + |
| 3a | 5 | + | + | + | - | - | - | + | + |
| 4a,b | 6 | + | + | + | + | - | + | + | + |
| 5c | 7 | + | + | + | - | - | - | - | - |
| 6 | 5 | - | + | + | - | + | - | + | + |
| 7 | 5 | - | + | + | - | - | - | - | + |
| 8d | 3.5 | + | + | + | - | - | - | - | - |
| 9e | 3 | - | + | - | + | Slightly | - | - | - |
| 10e | 6 | - | + | - | - | - | - | - | - |
| 11 | 4 | - | + | + | - | - | - | - | - |
| 12 | 2 | + | + | - | - | + | - | - | + |
| 13 | 2.5 | - | - | + | - | Slightly | - | - | - |
| 14 | 6 | - | Slightly | + | - | - | - | + | - |
| 15 | 2.5 | - | Slightly | - | - | - | - | - | + |
aMolecular confirmed cases.
bAtrial septal defect.
cHearing impairment.
dCongenital cataract.
eSkin hyperpigmentation.
5. Discussion
This study is the first Iranian study to evaluate the 11p15 methylation status in Russell-Silver syndrome patients. According to our results, 26.6% of the children had the confirmed results for Russell-Silver syndrome. It has been reported that up to 60% of the Russell-Silver cases will have a molecular cause while the molecular diagnosis in the rest of the patients will remain unknown. The loss of methylation on chromosome 11p15 has been reported as the most common underlying molecular etiology in Russell-Silver syndrome (8). Two imprinted domains in the 11p15 region have been associated with both Beckwith-Wiedemann syndrome and Russell-Silver syndrome. Hypomethylation of H19/IGF2 IG-DMR, which is a paternally methylated imprinting control region, will result in the reduced paternally expression of IGF2, as well as decreased maternal expression of H19 (5). These alterations will end up in growth restriction. In addition to these changes, the copy number variations in the 11p15 region have also been reported as a reason for the development of Russell-Silver syndrome. MS-MLPA and southern blotting are two main techniques in diagnosing 11p15 Russell-Silver syndrome (10). Another molecular cause of Russell-Silver syndrome has been located on chromosome 7, which was not evaluated in the present study. The upd(7)mat has been thought to be responsible for less than 10% of Russell-Silver syndrome cases (7, 11). Karaca et al. study evaluated upd(7)mat in a Turkish population and reported that 7.6% of the patients would have this defect (11). Moreover, Fuke et al. evaluated similar studies with a greater sample size on a Chinese population and find out upd(7)mat is responsible for 6.5% of the causes. They also provided a finding that H19-DMR alteration is responsible for 31,2% of the causes (12). Based on the most recent study performed on a greater population, the H19 loss of methylation and upd(7)mat were seen in 35.75 and 21.4% of the patients, respectively (13). In addition, 3.6% of the patients had upd(11)mat and around 40% had unknown causes. In addition to the mentioned molecular alteration, other abnormalities such as 14q32 abnormalities, upd(20)mat, upd(16)mat, and multilocus imprinting disturbance are other less common causes (5). The variety in molecular causes of Russell syndrome has made the molecular diagnosis complicated and expensive in centers that are not fully equipped for molecular studies. Therefore, most of these centers prefer the diagnosis only according to the clinical diagnostic criteria after excluding possible differential diagnosis. However, determining a clinical cutoff for Russell-Silver syndrome, as well as difficulties in performing molecular assessments, has made the diagnosis more complicated. Some studies have suggested patients who have scored three out of six on the Netchine-Harbison clinical scoring system may even have Russell-Silver syndrome, especially those with upd(7)mat (9). Although this syndrome can be presented with rare phenotypes (14), some researchers have tried to establish a relation between Russell-Silver phenotype and genotype. Wakeling et al. study is one of these studies that has evaluated those patients with 11p15 and upd(7)mat (15). Although they could not find any relation between disease severity and 11p15, they concluded that these patients are most likely to show classical Russell-Silver syndrome. Moreover, patients with upd(7)mat are most likely to show speech problems and learning difficulties. Although our study was conducted on a small group of Russell-Silver patients and we could not be able to conclude any phenotype-genotype correlation, the most common findings in our 15 patients were triangular face and feeding difficulties.
5.1. Study Limitation
According to our limitation in time and resources and its rare disorder, we were not able to include more children in our study. Moreover, the financial cost of performing further evaluations for other causes of Russell-Silver syndrome such as upd(7)mat was another limiting factor in our study.
5.2. Conclusions
Russell-Silver syndrome is a rare genetic syndrome that has not been widely studied in Iran. This syndrome as one of the primordial dwarfism syndromes requires more attention in term of molecular detection. According to our results, only 26.6% of our study population had confirmed MLPA results, which is lower than the reported studies. Further studies for determining other molecular causes of this syndrome are warranted.