1. Background
It has been estimated that about 95% of Congenital Adrenal Hyperplasias is produced by a mutation in the CYP21A2 (cytochrome p450 family 21 subfamily A member 2) gene. The CYP21A2 gene contains 10 exons, about 3500 bp and encode steroid 21-hydroxylase (21-OHD) protein which is a member of the cytochrome p450 super-family enzymes (1). The occurrence of the classical form in the Caucasian population is 1:10,000 - 1:15,000 (2), while the occurrence of the non-classic CAH form is significantly higher and more variant from 1:1000 to 1:2000 (3).
CYP21A2 gene is situated inside the HLA class III (human histocompatibility complex) locus on chromosome 6p21.3 region. CYP21A2 along with its adjacent genes (RP1, C4, TNXB, and their truncated pseudogenes RP2 and TNXA) produce a genetic unit called the RCCX module (RP-C4-CYP21-TNX) which generates an extremely variable region of approximately 30 kb of DNA (4, 5).
Originally, there are two common classifications and three known forms of CAH. Individuals with the salt-wasting (SW) type, have CYP21A2 gene mutations which resulted in a completely loss of 21-OHD enzyme function. While those with the simple virilizing (SV) type, release only a low levels of the functional enzyme. It is noticeable that about 1% to 2% of enzyme activity is enough to make some aldosterone so they do not have problems with salt wasting.
In non-classical form of CAH, the amount and also the activity of the enzyme is reduced, though the enzyme activity is more in comparison to the other types (6). The mutations which lead to milder form of the disease retain 20 to 50% of the enzyme activity (7). This form of CAH may manifest with signs and symptoms of androgen excess or may be asymptomatic (8).
In some cases, a single mutation leads to a single recognized clinical form but in many other cases multiple mutations lead to this phenotype as in CAH, where more than 70% of it are heterozygote compounds (7, 9, 10).
It is noteworthy that more than 100 mutations have been found in the CYP21A2 gene which can cause 21-hydroxylase deficiency (2). Some of these mutations result from an exchange of genetic material between the CYP21A2 gene and a comparable but nonfunctional piece of DNA called a CYP21A1P pseudogene, which is located 30kb upstream to the CYP21A2 gene on chromosome 6. The gene and pseudogene share 98% sequence homology in exons and 96% in the non-coding region (11). The type of DNA alternation is considered as an unequal crossing over. Other mutations which are involved in 21-OHD deficiency include point mutations and large deletions or insertions of the CYP21A2 gene (12).
2. Objectives
The purpose of our study was to describe a comprehensive CYP21A2 mutation analysis in a cohort of CAH patients and determine the frequency of genetic variations that complicate the accuracy of routine genotyping of patients of our country. Also, for the first time we report the different polymorphisms of the CYP21A2 gene in the Iranian population.
3. Methods
3.1. Patients
The current study was conducted on 25 Iranian CAH patients (6 females and 19 males) with twenty-two unrelated families whose ages ranged from 6 months to 21 years. Fourteen families had consanguineous marriage. The informed consent for participation in the study was obtained from patients or their parents. Blood samples were collected of patients referred to Ali-Asghar Hospital in Tehran. All patients were clinically diagnosed as CAH. They originated from different regions of Iran. This is an experimental study.
3.2. PCR Amplification
Genomic DNA was isolated from peripheral blood using Yekta Tajhiz Azma (YTA) kit (Iran). CYP21A2/CYP21A1P chimeras, resulted from the unequal crossing over were analyzed using two PCRs, by sequence- specific primers (2-7, 9-13).
Initially, to detect the normal gene from pseudogene, PCR was performed by employing two specific sets (Table 1). To recognize the presence of a recombination site, a fragment of 1327 bp spanning from exon 1-6 was amplified using a CYP21-specific forward (F1) and CYP21-specific reverse (R1) primer. CYP21P-specific forward primer (F1/P1) with the same reverse primer of the gene (R1) was used to amplify the pseudogene. Similarly, a second PCR reaction was set up. A CYP21P-specific (F2) or CYP21-specific (F2/P2) forward primer was used in combination with CYP21-specific reverse primer (R2) to detect either the allele with a recombination in this region or the normal allele (exon 6-10) respectively, and a fragment of size 1800 bp was amplified (Table 1). The PCR program exists in Appendix 1 in Supplementary File.
| PCR Set/Primers | Position | Sequence (5’ → 3’) | Product Size, bp |
|---|---|---|---|
| PCR set1 | 1327 | ||
| F1 (forward) | EXON1, Normal gene | 5'-TCCGGAGCCTCCACCTCCC-3' | |
| F1/P1 (forward) | EXON1, pseudogene | 5'-TCCGGAGCCTCCACCTCCT-3' | |
| R1 (reverse) | EXON 6, Normal gene | 5'-AGCTGCATCTCCACGATGTGA-3' | |
| PCR set2 | 1800 | ||
| F2 (forward) | EXON 6, Normal gene | 5'-GCCATAGAGAAGAGGGATCACAT-3' | |
| F2/P2 (forward) | EXON 6, pseudogene | 5'-GCCATAGAGAAGAGGGACCACAA-3' | |
| R2 (reverse) | EXON10, Normal gene | 5'-TTAAGCCTCAATCCTCTGCAGCG-3' |
After separating gene from pseudogene, in a second step, PCR performed on 19 patients, which followed by sequencing of gene to distinguish the mutations in main gene. In order to analyze the 1 - 7 exons of CYP21A2, we used three different primers which were unable to amplify pseudogene. Primer sequences and PCR amplification are described in supplementary file appendix 1 (13). Furthermore, the PCR products of exon 6 - 10 (if did not have chimeras) were sequenced with set2 primers (Table 1).
Sequencing was carried out by MACROGEN company in South-Korea, using the classic Sanger method with ABI. Data analysis was performed by Sequence Scanner v1.0 and Chromas software.
4. Results
In the initial scanning, the data indicated that 6 (24%) probands had unequal crossing over (CYP21A1P/CYP21A2 chimeras). The remaining 19 (76%) patients had a point or compound heterozygous mutations or deletion in the CYP21A2 gene (Table 2).
| Genotype | Phenotype | Number of Patients | Frequency, % |
|---|---|---|---|
| p.I173N/exon 6 cluster/ p.V282L | SV | 9 | 47.3 |
| IVS2-13A>C,G/ exon 6 cluster/p.V282L | SW | 1 | 5.2 |
| p.I173N/exon 6 cluster | SV | 3 | 15.7 |
| p.G110Efs/ p.G110Efs | SW | 1 | 5.2 |
| p.I173N/- | SV | 2 | 10.5 |
| Exon 6 cluster/- | SV | 2 | 10.5 |
| Exon 6 cluster/ p.V282L | SV | 1 | 5.2 |
Overall among 25 recognized patients, 16 (64%) possessed the SV-CAH form and 9 (36%) had the SW-CAH form.
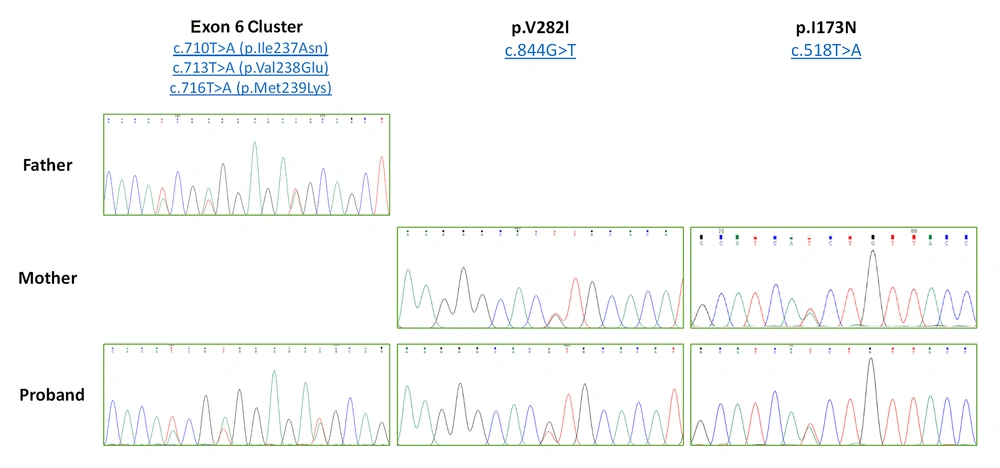
In our study, p.I173N/exon 6 cluster/p.V282L compound heterozygous mutations were the most frequent (47.3%) ones (Figure 1).
Electropherogram of compound heterozygous mutation (exon 6 cluster/p.V282L/p.I173N). There is exon 6 cluster in father and p.V282L/p.I173N variant in mother as heterozygote form. In proband graph, all three variants (exon 6 cluster, p.V282L and p.I173N) exist together causing pathogenic compound heterozygote form. The 8 nucleotide deletions in exon 3 and IVS2-13A>C, G/exon 6 cluster/p.V282L and Exon 6 cluster/p.V282L as compound heterozygous mutations were less frequent (5.2%). The frequency of the mutated alleles in patients is shown in Table 2.
To analyze genotype-phenotype correlations, disease-causing mutations and polymorphisms were divided into four groups according to Finkielstain et al. (14). Our patients were phenotypically placed in the same group as their patients which confirms the existence of genotype-phenotype correlations. In summary, Group 0, included patients with null mutations (gene deletions, p.G110Efs; exon 6 cluster) on both alleles (no enzymatic activity). Group A, included patients with compound heterozygous for IVS2-13A/C>G. IVS2-13A/C>G, which is known to have minimal residual enzymatic activity. Group B, included patients with p.I173N (2% residual enzymatic activity) and any other homozygous or compound heterozygous mutations that exist in groups 0, or A. Group C, included patients with p.V282L mutations (20% - 60% residual enzymatic activity) compound heterozygous with groups 0, and A or B (Table 3).
| Patient | Genotype Group | Gender | Pathogenic Mutations | Polymorphisms | Phenotype |
|---|---|---|---|---|---|
| 1 | B | Male | p.I173N/exon 6 cluster/p.V282L | Rs6468 | SV |
| 2 | A | Female | IVS2-13A/C>G /p.I173N/exon 6 cluster | - | SW |
| 3 | B | Male | Exon 6 cluster/p.I173N | Rs6474-rs6477, Rs6472-rs778403992, c.29-31delCTG | SV |
| 4 | B | Male | p.I173N /exon 6 cluster/p.V282L | Rs6477 | SV |
| 5 | B | Male | p.I173N/- | Rs530758070-rs6477, c.29-31delCTG | SV |
| 6 | B | Male | Exon 6 cluster/p.I173N | Rs530758070-rs6477 | SV |
| 7 | B | Female | Exon 6 cluster/p.I173N | Rs6474-rs6446-rs6473-rs562025438 | SV |
| 8 | B | Male | p.I173N/exon 6 cluster/p.V282L | Rs11970671 | SV |
| 9 | B | Female | p.I173N/exon 6 cluster/p.V282L | Rs6477 | SV |
| 10 | 0 | Male | Exon 6 cluster/- | RS6477 | SV |
| 11 | B | Male | p.I173N/- | - | SV |
| 12 | B | Male | p.I173N /exon 6 cluster/p.V282L | c.29-31delCTG | SV |
| 13 | B | Female | p.I173N /exon 6 cluster/p.V282L | Rs6477 | SV |
| 14 | 0 | Male | Exon 6 cluster/- | Rs6472-c.29-31delCTG | SW |
| 15 | B | Male | p.I173N /exon 6 cluster/p.V282L | - | SV |
| 16 | 0 | Male | G110Δ8nt/ G110Δ8nt | - | SW |
| 17 | B | Female | p.I173N /exon 6 cluster/p.V282L | - | SV |
| 18 | C | Male | Exon 6 cluster/ p.V282L | Rs6472-Rs6473-rs6446-c.29-31delCTG-Rs11970671 | SV |
| 19 | B | Male | p.I173N /exon 6 cluster/p.V282L | Rs6473-rs6446-rs6477-c.29-31delCTG | SV |
The results were evaluated by considering the international genetic databases “dbSNP” and “The Human Gene Mutation Database” HGMD”. Variations were found in 14 patients as follows: The variants rs6477 [c.747C>G(p.Leu249=)], rs6474 [c.118C>T(p.Leu40=)], rs6446 [c.1473G>A(p.Pro491=)], rs1058152, rs562025438 [c.*39G>A], rs11970671 [c.822T>C(p.Ser274=)], rs530758070, rs6468 [c.118C>T(p.Leu40=)], rs6472 [c.806G>C(p.Ser269Thr)], rs6473 [c.1481G>A(p.Ser494Asn)] and rs193922546 [c.342C>T (p.Ser114=)] found in this study were reported as “polymorphisms” in the dbSNP and HGMD (Table 4).
| Number of SNP | Exon | Protein | Patient Genotype (No.) | SNP code | ||
|---|---|---|---|---|---|---|
| 1 | Exon 1 | p.Leu40= | C/T (2) | C/C (0) | T/T (0) | rs6468 |
| 2 | Exon 3 | p.Arg103Lys | A/G (4) | A/A (0) | G/G (0) | rs6474 |
| 3 | Exon 3 | p.Ser114= | C/T (1) | C/C (0) | T/T (0) | rs193922546 |
| 5 | Exon 7 | p.Leu249= | C/G (13) | C/C (0) | G/G (1) | rs6477 |
| 6 | Exon 7 | p.Ser269Thr | C/G (4) | G/G (0) | C/C (0) | rs6472 |
| 7 | Exon 7 | p.Ser274= | C/T (1) | C/C (0) | T/T (0) | rs11970671 |
| 8 | Exon 10 | p.Pro491= | A/G (4) | A/A (0) | G/G (0) | rs6446 |
| 9 | Exon 10 | p.Ser494Asn | A/G (4) | A/A (0) | G/G (0) | rs6473 |
| 10 | 3’UTR | unknown | C/T (1) | C/C (0) | T/T (0) | rs1058152 |
| 11 | 3’UTR | c.*39G>A | C/T (1) | C/C (0) | T/T (0) | rs562025438 |
5. Discussion
In CAH, mutations are usually categorized in compound heterozygous status, which possess diverse 21-OHD enzymatic activity. Therefore, there is a high degree of phenotypic overlapping between moderate and mild forms of CAH, reflected by the wide and heterogeneous spectrum of clinical manifestations. The CYP21A2 gene variations determined in the patients, supported the clinical signs of the disease (8, 12, 15, 16).
In previous reports, about 65% - 75% of CAH patients that had different mutations of the CYP21A2 gene were in compound heterozygous state. This study sequenced the whole gene including introns and the data showed that majority (68%) of patients were compound heterozygotes which is compatible with other reports around the world (17, 18). In a previous study of CAH in Iran, by Forouzanfar et al., 2015, 13 of 21 (61%) patients had compound heterozygote mutations which is close to our study, although in Rabbani et al. 2008 study, 26 cases of 44 (59%) patients had compound heterozygote mutations (19), a rate was less than ours (68%).
Our study showed 6 (24%) patients had chimeric gene mutation. The most frequent (31.8%) mutations detected by Rabbani et al. in 2008, were gene deletions and chimera (10, 20) which is close to our results. It is stated that pseudogenes could be exposed to crossing over and chimeric mutation (21).
Some studies declare more than one mutation in some alleles of CYP21A2 gene (14, 19, 20, 22-24). Finkielstain et al. (14) found some cases with classic form of CAH which had complex alleles comprising 3 mutations (p.I173/N+ other mutations: exon 6 cluster, p.V282L, p.Leu307fs, p.Q318X). Similarly, we found three allelic compound heterozygote mutations in 8 patients with SV-CAH form. However, in one case, a sixteen-year old girl with a multiplex compound heterozygous pathogenic mutation (I2G/exon 6 cluster, p.V282L) we found it in SW form. These findings are consistent with the studies mentioned above.
Compound heterozygote mutations of exon 6 cluster (I236N-V237E-M239K)/p.V282L/ p.I173N were found in 9 patients. In order to confirm the accuracy of mutations, their parents were also analyzed separately. In each family, one of the parents had a heterozygote variant (p.I173N/p.V282L) and other one had exon 6 cluster heterozygote variant without any symptoms of the disease. Of course, existence of these three variants (exon 6 cluster, p.V282L and p.I173N) in the heterozygous form in each individual are pathogenic but it seems that in our population these two variants (p.I173N/p.V282L) and existence of exon 6 cluster by itself is not considered as pathogenic. It should be considered that each of these variants by themselves are pathogenic in the homozygote form (25).
Association between exon 6 cluster and p.V282L, seems not pathogenic in our study as parents and their children had both mutations in heterozygote form, and we did not find this compound heterozygote in reviewed papers as pathogenic, but in one study of Bas et al. (23, 26, 27) p.I173N/exon 6 cluster compound heterozygote mutation was seen in three patients. This mutation was previously reported in the study of Finkielstain et al. as compound heterozygote with SV form. Our patients showed the same clinical symptoms (14).
G110Δ8nt mutation is an 8 bp deletion in exon3 that creates SW-CAH form and was identified in one of our patients. The frequency of this mutation was previously reported as 4.5% in Iran (20) and 4.8%, 4.3% and3.2% in USA (28), Netherlands (29) and Iraq (30), respectively. Our finding (4%) was compatible with that of other researches.
The more interesting finding of our study is submission the percent of variants in CYP21A2 gene, for instance: rs6477 (56%), rs6468 (8%), rs6474 (12%), rs6472 (16%), rs6473 (16%), rs6446 (16%) andrs193922546, rs530758070, rs11970671, rs61732108, rs778403992, rs1058152 and rs562025438 each (4%). The current study is the first report which gives distribution of the variants in Iranian population; however it was reported in other countries of the world (31, 32). Additionally, our study indicated correlation between some of these variants, which is shown in Table 4. A study conducted in 2015 by Gürkan et al. confirms this correlation (31).
It has been shown that rs6467 (HGMD: CS880069, rs6472HGMD: CM994664 and rs6473 have been associated with classical CAH (21-hydroxylase deficiency) and rs6445 has been associated with CAH (21-hydroxylase deficiency) non-classical type. rs6474, rs6477 and rs61338903(c.29-31delCTG) belong to a non-pathogenic variant and are without effects on the 21-OHD activity according to HGMD and dbSNP (31). Also, in our findings, this association between variants and phenotypes is consistent with HGMD and dbSNP (Table 4).
Gurgov et al. examined the association between CYP21A2 mutations and virilization, and showed either one of the following mutations: (A) deletion, (B) I2G, and (C) I173N combined, demonstrate ambiguous genitalia in 94% of case. This confirms high reliability of predicting the degree of virilization based on the genotype in these groups. In our study, when I173N was combined with any other mutation in compound heterozygote form, virilization was seen, too (25).
Indeed, we reported a heterozygote variant c.29-31delCTG (rs61338903) that was detected in 8 patients and there was a deletion of a CTG repeat in tandem repeats of 5 CTGs. Rodrigues et al. have reported an insertion in this position, but this insertion had no effect on the enzymatic activity (33).
5.1. Conclusions
In conclusion, a great diversity of compound heterozygosity was found in our population with large spectrum of mutated alleles. Therefore, The PCR-sequencing of the whole gene (CYP21A2) seems a preferred method, although the first step approach could be analyzing CAH patients based on the prevalence of the mutations of this gene in case that there is limitations of funding. Our patient’s clinical manifestations were correlated with mutated alleles and the residual activity of 21-hydroxylase enzyme. This study could help genetic counseling of these patients in our population and contribute to better understanding of the molecular landscape of CAH in Iran.
Acknowledgment: