1. Background
Brucellosis is a zoonotic disease that infects about 500,000 people annually (1). This disease has been observed in many geographical regions, including the Mediterranean, Middle East, Central and Southern America, Africa, and Asia (2). Brucellosis can be transmitted through the secretions of infected animals, but the consumption of unpasteurized dairy products, such as goat milk and fresh cheese, is the most common cause of infection. For humans, antibiotic therapy is recommended to treat brucellosis. In contrast, infected animals are often slaughtered due to a lack of drug adaptation and affordability (3).
To prevent brucellosis in animals, various strategies are employed, including inoculation, adherence to sanitary principles, and detection and elimination of infected animals. In general, attenuated vaccines are used to control brucellosis in humans. Although these vaccines have had some success, their application has been limited due to low efficiency and the issue of antibiotic resistance. Therefore, given the sanitary and economic importance of brucellosis, along with serious problems in its treatment and control, novel and precise methods should be investigated.
An efficient vaccine used to control brucellosis should have several advantages, including stability in the host body, the absence of disease triggering in the host, prevention of abortion, and the capacity for large-scale production (4). Currently, recombinant vaccines (RVs) employing different antigenic proteins are recommended as an intriguing method to control various infectious diseases (5). These vaccines not only have all the advantages of an efficient vaccine but can also be flexibly designed for specific applications. Despite the advantages of RVs, low antigenicity is known as their most significant disadvantage; consequently, a molecular adjuvant should be conjugated with RVs to enhance their immune responses (6).
In general, outer membrane proteins (OMPs) are known as immunogenic proteins in Brucella that can strongly stimulate immune responses (1). Different members of OMPs have been evaluated for Brucella; among these proteins, outer membrane protein 25 (OMP25) has been strongly recommended for RV design due to its protective properties (7). Mycobacterium tuberculosis heparin-binding hemagglutinin adhesin (HBHA) has been extensively applied as a molecular adjuvant for designing RVs (6, 8). This protein, as a novel antagonist of the toll-like receptor 4 (TLR4) receptor without toxicity in the host body, can play a role in the maturation of dendritic cells and enhancement of immune responses (9).
2. Objectives
The present study was conducted to design and engineer an RV based on the OMP25 protein of Brucella (as an antigenic moiety) and the HBHA protein of M. tuberculosis (as a molecular adjuvant moiety).
3. Methods
3.1. Protein Achievement
To carry out the current project, the amino acid sequences of OMP25 and HBHA proteins, with accession numbers JF918759.1 and NC-000962.39 (respectively), were obtained from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). Additionally, the PDB file of the TLR4/MD2 receptor, with an accession number of 2z64, was obtained from the Protein Data Bank (https://www.rcsb.org/).
3.2. Engineering a Chimera of HBHA-OMP25
To engineer a chimera of HBHA-OMP25, CLC Main WorkBench software version 5.2 was employed. In this process, the first signal peptide sequences of both proteins (HBHA and OMP25) were removed, and the correction of the reading frames was evaluated. Finally, the amino acid sequence of the HBHA protein (at the N terminal) was linked to the amino acid sequence of the OMP25 protein (at the C terminal) using an appropriate rigid linker.
3.3. Investigation of Important Features
To investigate the physical and chemical features (e.g., protein weight, GRAVY, pI, stability indicator, and aliphatic indicator) of the HBHA-OMP25 chimera, the ProtParam tool was used. Additionally, the VaxiJen tool with a cutoff of 0.5 was employed to evaluate the antigenicity of the designed construct, while its allergenicity was investigated using the AllerTOP tool.
3.4. Investigation of Different Structures
To investigate the secondary structure of the chimera, the SOPMA tool was applied. In this case, the amino acid sequence of the chimera was uploaded to the tool. Additionally, to predict the three-dimensional (3D) structure of the chimera, the I-TASSER tool was used. Refinement of the 3D structure was conducted using the GalaxyRefine tool, and the best-refined model was chosen based on the outputs from the VADAR tool.
3.5. Protein-Protein Docking
To investigate the docking between the HBHA moiety of the chimera and the TLR4/MD2 receptor, a protein-protein docking analysis was performed using the ClusPro tool. For this purpose, the PDB format of the best-refined model of the chimera and the PDB file of the TLR4/MD2 receptor were uploaded to the tool. Additionally, the hydrogen bonds of the complex were analyzed using LigPlot+ software.
3.6. Codon Adaptation and Computer-Aided Cloning
In this project, codon adaptation of the chimera nucleotide sequence for expression in a prokaryotic system was performed using the JCat tool (http://www.jcat.de/). The JCat tool employs a precise algorithm to predict the codon adaptation index (CAI), as presented by Carbone et al. (10). This algorithm can prepare a reference set of genes that are highly representative of the bias. Finally, the cloning potential of the adapted sequence into pET21a (+) was verified. In this process, BamHI and NotI restriction enzyme sites were added to the 5' and 3' regions of the adapted sequence.
4. Results
4.1. Engineering a Chimera of HBHA-OMP25
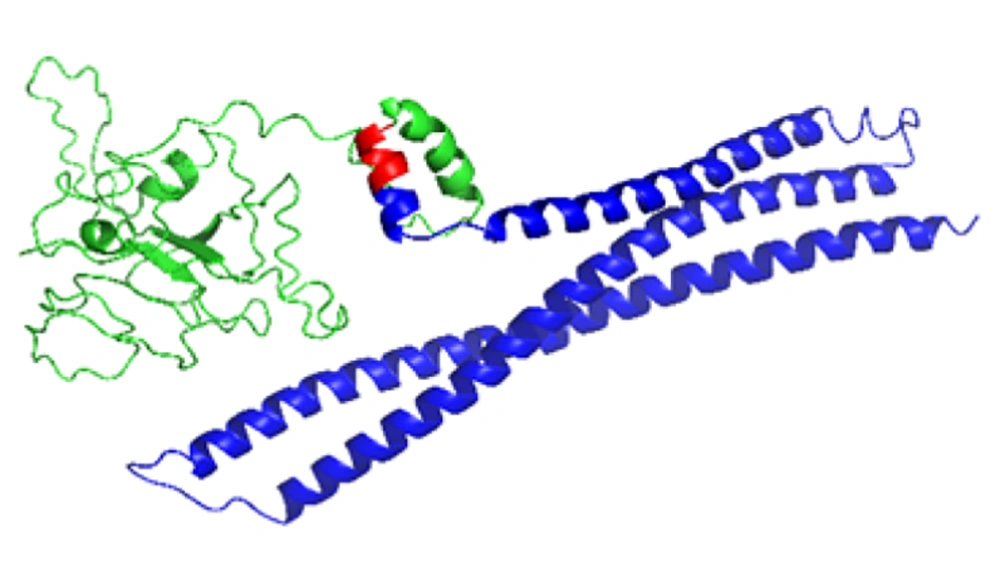
The engineered chimera, from the N-terminal to the C-terminal, contained HBHA and OMP25 moieties, respectively (Figure 1). Additionally, an EAAAK linker, consisting of five amino acid residues, was embedded between the two moieties (Figure 1). It must be noted that the entire designed chimera contained 417 amino acid residues.
Final construct of the engineered chimera which was designed by CLC WorkBench software. Molecular adjuvant [heparin-binding hemagglutinin adhesin (HBHA)] moiety, rigid linker (EAAAK) and antigenic moiety [outer membrane protein 25 (OMP25)] have been illustrated by green, red and blue, respectively.
4.2. Investigation of Important Features
Based on the physical and chemical results, the protein weight, pI, aliphatic index, instability index, and GRAVY of the chimera were 45.172 kDa, 9.03, 80.46, 34.95, and -0.450, respectively. Additionally, the half-life of the engineered chimera in mammals, yeast, and E. coli was 30 hours, > 20 hours, and > 10 hours, respectively. According to the results, the antigenicity score of the engineered chimera was 0.7184, classifying it as an antigen, while the allergenicity results confirmed that the engineered chimera was a non-allergenic protein.
4.3. Investigation of Different Structures
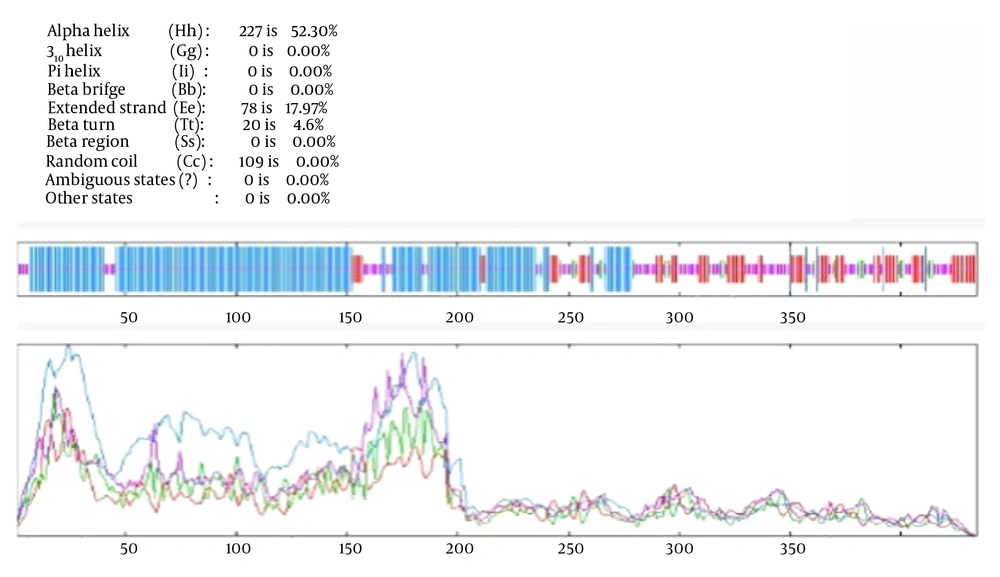
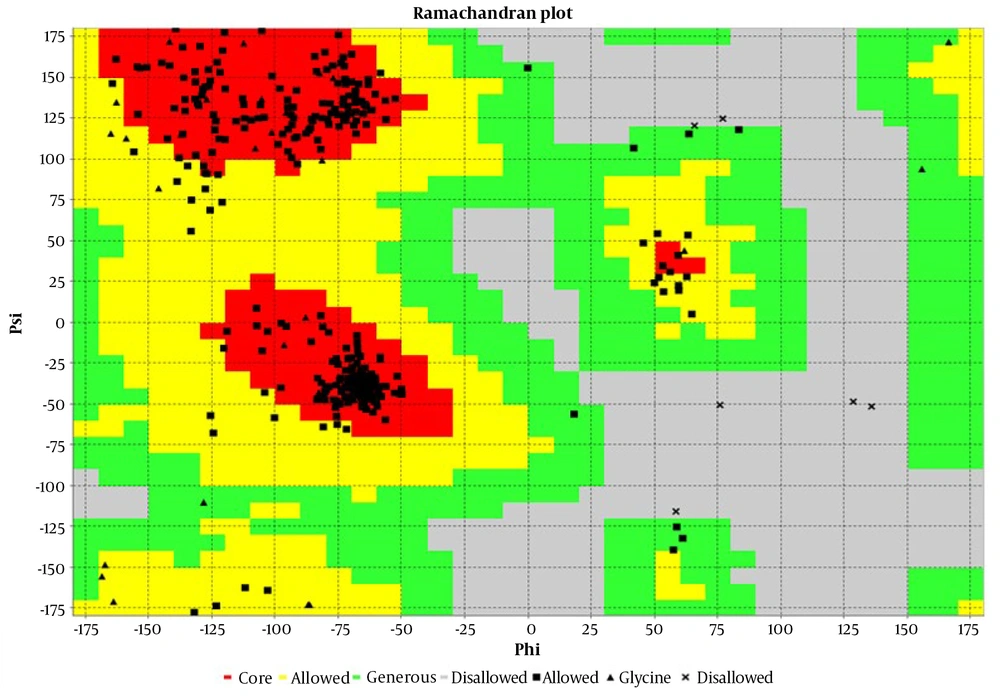
As mentioned, to predict the secondary structure of the engineered chimera, the SOPMA tool was used. The results demonstrated that the engineered chimera contained 52.30% alpha helix, 17.97% extended strand, 4.61% beta turn, and 25.12% random coil (Figure 2). The 3D structure of the engineered chimera was predicted by the I-TASSER tool with a C-score of -1.32 (Figure 3). Furthermore, the Ramachandran analysis confirmed that 89% of the amino acid residues of the most refined model were in the core region, while only 1% were in the disallowed region (Figure 4).
4.4. Protein-Protein Docking
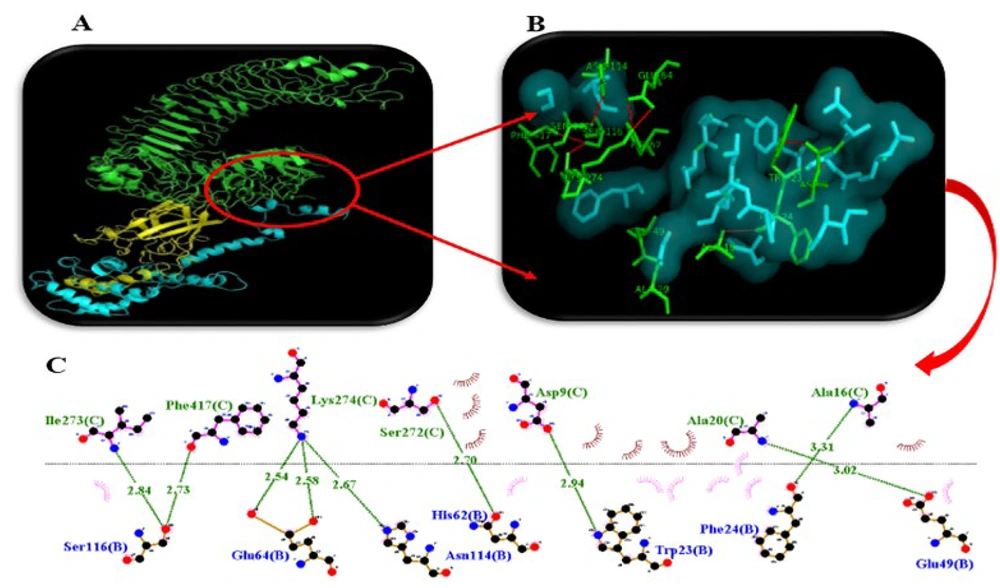
As mentioned, in the current project, a protein-protein docking strategy was employed to investigate the interaction between the HBHA moiety of the engineered chimera and the TLR4/MD2 receptor. The results confirmed that the HBHA moiety could successfully dock with the TLR4/MD2 receptor with a lowest energy of -0.777 kcal/mol (Figure 5A). Additionally, the results from LigPlot+ software revealed that nine hydrogen bonds were formed in this interaction (Figure 5B).
Interaction between the chimera of heparin-binding hemagglutinin adhesin (HBHA)- outer membrane protein 25 (OMP25) and toll-like receptor 4 (TLR4)/MD2 was visualized by PyMol software. A, protein-protein docking between the HBHA moiety (yellow) and the TLR4/MD2 receptor (green); B, amino acids involved in the interaction (green) between the HBHA moiety and TLR4/MD2 receptor; C, hydrogen bonds (green lines) formed between the HBHA moiety and TLR4/MD2 receptor.
4.5. Codon Adaptation and Computer-Aided Cloning
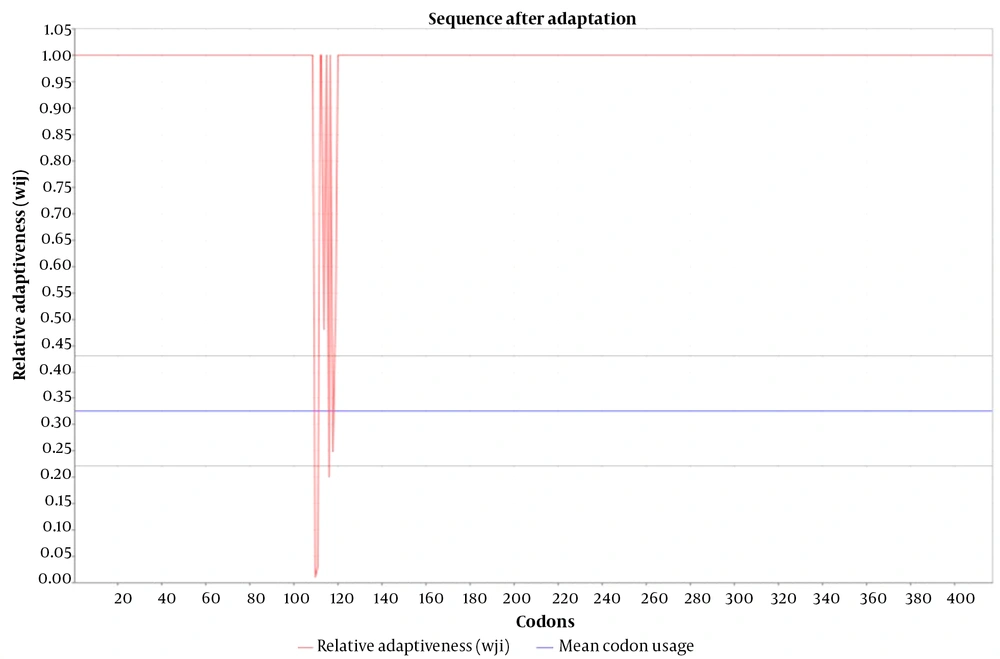
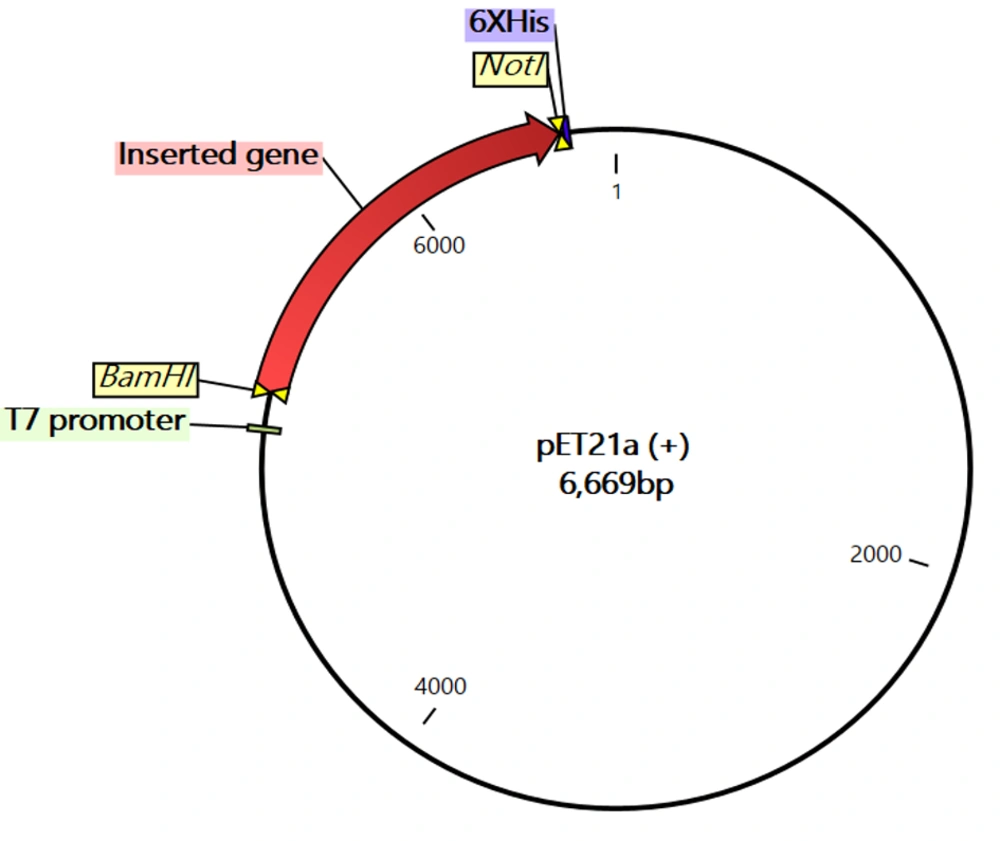
To perform codon adaptation of the nucleotide sequence of the engineered chimera, the JCat tool was used. The results of this analysis demonstrated that the CAI of the sequence before and after adaptation were 0.375 and 0.970, respectively, whereas the GC contents of the sequence before and after adaptation were 61.9% and 51.8%, respectively (Figure 6). Finally, the results of computer-aided cloning revealed that the nucleotide sequence of the engineered chimera could be correctly cloned between the BamHI and NotI enzymes of the multiple cloning sites of the pET21a (+) vector (Figure 7).
Computer-aided cloning of the engineered chimera nucleotide sequence. The inserted gene (red region) with 1258 nucleotides in length was cloned between BamHI and NotI enzymes at 5' and 3' regions, respectively. The most important regions of the vector including T7 promoter and His tag have been shown with green and purple, respectively.
5. Discussion
Brucellosis is a zoonotic disease prevalent in most developing countries (11). Despite livestock vaccination efforts, brucellosis continues to cause significant economic damages and human infections (12-14). Currently, antibiotic therapy is used to treat brucellosis; however, this method is not entirely efficient (13). In contrast, a vaccination strategy proves to be more effective and affordable (13, 14). Therefore, vaccine development is strongly recommended to eradicate brucellosis. Since the beginning of the 20th century, numerous studies have been conducted to develop an ideal vaccine for brucellosis (15). So far, different types of vaccines, such as live and inactivated vaccines, have been used to control brucellosis. Despite their success, these vaccines have several disadvantages (16).
Currently, a third generation of vaccines, which combines bioinformatics and genetics, is being developed. This generation leads to the production of safe vaccines that only use the immunogenic components of a pathogen (17, 18). The current project aimed to engineer and develop an HBHA-OMP25 vaccine against brucellosis using bioinformatics. In this case, the OMP25 protein of Brucella melitensis was considered as an immunogenic protein. Outer membrane protein 25 is an outer membrane protein with a molecular weight of 25 kDa found in all species of Brucella (4). It has been reported that OMP25 plays an important role in triggering the pathogenicity and viability of B. melitensis in its host (19). Bowden et al. (20) and Commander et al. (21) reported that using OMP25 as a recombinant protein could lead to resistance against brucellosis in mice. Moreover, Yousefi et al. (19) confirmed that OMP25 can be used as an effective vaccine candidate against brucellosis.
In the current study, besides OMP25, the HBHA protein was used as an adjuvant to enhance the potency of the engineered chimera. The HBHA protein is a TLR4 receptor antagonist known to strengthen immune responses, making it useful as a molecular adjuvant (22). As mentioned in this study, the EAAAK linker was used to assemble the HBHA-OMP25 chimera. Fusion protein linkers play a key role in maintaining the functions of chimeric proteins. EAAAK is a rigid linker that preserves the functionality of different moieties within a chimeric protein (23).
In continuation, the physical and chemical features of the engineered chimera were investigated. Our analysis revealed that the molecular weight of the protein was 45.172 kDa. It has been confirmed that proteins with a molecular weight greater than 10 kDa can evade the renal system of the kidney, consequently increasing their half-life in the host (17). The instability index is an important parameter related to the stability of a protein. In general, a protein is considered stable when the instability index is less than 40, whereas proteins with an index greater than 40 are considered unstable. Our results showed that the instability index of our engineered chimera was 34.95, indicating it is a stable protein (24, 25).
Additionally, antigenicity results showed that our engineered chimera, with an antigenicity score of 0.7184, could stimulate the immune system. The 3D structure analysis of our engineered chimera revealed that 89% of amino acid residues were located in the core region, indicating appropriate 3D structure modeling. Our protein-protein docking results demonstrated that the HBHA moiety of the engineered chimera could interact with the TLR4/MD2 receptor with the lowest energy of -0.777 kcal/mol and nine hydrogen bonds, as shown in Table 1. To express our engineered chimera in a prokaryotic expression system, the codons of its nucleotide sequence were adapted for this system. Our results showed that the CAI (the most important indicator) of the adapted sequence improved from 0.375 to 0.970. It must be noted that the maximum value for CAI is 1; therefore, when CAI reaches 1, all codons are fully adapted (10).
| HBHA Moiety | TLR4/MD2 Receptor | Length of the Bond |
|---|---|---|
| Isoleucine 273 | Serine 116 | 2.84 |
| Phenylalanine 417 | Serine 116 | 2.73 |
| Lysine 274 | Glutamic acid 64 | 2.54 |
| Lysine 274 | Glutamic acid 64 | 2.58 |
| Lysine 274 | Asparagine 114 | 2.67 |
| Serine 272 | Histidine 164 | 2.70 |
| Aspartic acid 9 | Tryptophan 23 | 2.94 |
| Alanine 20 | Glutamic acid 49 | 3.02 |
| Alanine 16 | Phenylalanine 24 | 3.31 |
List of Amino Acid Residues Involved in Hydrogen Bonds of Heparin-Binding Hemagglutinin Adhesin and Toll-Like Receptor/MD2 Interaction
5.1. Conclusions
In this study, based on the OMP25 of B. melitensis and the HBHA protein (as a molecular adjuvant), a novel chimera of HBHA-OMP25 was designed. Our various results confirmed that the designed chimera could not only be an appropriate RV against brucellosis but also be easily produced using a prokaryotic expression system.
![Final construct of the engineered chimera which was designed by CLC WorkBench software. Molecular adjuvant [heparin-binding hemagglutinin adhesin (HBHA)] moiety, rigid linker (EAAAK) and antigenic moiety [outer membrane protein 25 (OMP25)] have been illustrated by green, red and blue, respectively. Final construct of the engineered chimera which was designed by CLC WorkBench software. Molecular adjuvant [heparin-binding hemagglutinin adhesin (HBHA)] moiety, rigid linker (EAAAK) and antigenic moiety [outer membrane protein 25 (OMP25)] have been illustrated by green, red and blue, respectively.](https://services.brieflands.com/cdn/serve/3170b/890834c725525cbd158410ed265501f0d647d934/JID-150899-i001-F1-preview.webp)