1. Background
Systemic lupus erythematosus (SLE) is a multisystem autoimmune disease characterized by the production of autoantibodies, hyperactivity of T and B cells, immune complex deposition, and damage to multiple organs. Patients with SLE can experience a range of systemic manifestations and organ-specific injuries. Additionally, they may experience nonspecific symptoms such as fatigue, fever, arthralgia, malaise, headache, and weight loss, which are general manifestations that can mimic other diseases (1, 2). Approximately half of lupus patients develop more severe complications, including nephritis, central nervous system vasculitis, pulmonary hypertension, interstitial lung disease, and stroke.
Several factors, including hormones, pathogens, medications, and UV radiation, are considered triggers and aggravators of SLE (3). Epidemiological studies have also indicated a significant contribution of genetic and epigenetic factors to the etiology of SLE. This disease shows high heritability (43 - 66%), with concordance rates of 24 - 57% in monozygotic twins and 2 - 5% in dizygotic twins or siblings (4-6). The prevalence of SLE varies by gender and ethnic background. The ratio of women to men is 9: 1, with a peak incidence in women during their reproductive years. SLE has been shown to be more frequent among African Americans and individuals of non-European descent (7, 8). In the Iranian population, the reported incidence of SLE is 40 per 100,000 people. Iranian patients tend to exhibit more severe symptoms than European Caucasians, which may be associated with risk factors such as ethnicity and diet (9).
The programmed cell death 1 (PDCD1) gene has been identified as a potential risk gene for SLE (10). Programmed cell death 1 (also known as PD1) encodes a member of the immunoglobulin superfamily that contains an immunoreceptor tyrosine-based inhibitory motif (ITIM). During cell activation, the tyrosine residue of the ITIM motif is phosphorylated, leading to the deactivation of downstream signaling molecules. This inactivation inhibits cytokine production and halts cell proliferation in the G1 phase of the cell cycle. Programmed cell death 1 is crucial for maintaining peripheral immunological tolerance by suppressing or inactivating self-reactive T and B cells, thereby preventing autoimmunity (11, 12). The PD-1.5 C/T polymorphism is located within exon 5 of the gene, and because it is a silent mutation, the final amino acid sequence of the protein remains unchanged. However, this single nucleotide polymorphism (SNP) could have a direct or indirect functional effect on PD-1.
It is important to investigate the relationship between PDCD1 gene polymorphism and SLE susceptibility in Iranians due to racial and regional differences. This research could enhance our understanding of the molecular basis and pathophysiology of SLE.
2. Objectives
This study aimed to investigate the association between the PD-1.5 C/T (rs2227981) SNP and susceptibility to SLE in a sample population from northwestern Iran.
3. Methods
3.1. Patients and DNA Samples
This case-control study included 52 unrelated patients and 53 healthy controls with no history of autoimmune abnormalities or cancer. The inclusion criteria for the case samples were a confirmed diagnosis of SLE by a specialist according to the American College of Rheumatology (ACR) classification criteria (Hochberg, 1997). Participants were selected from northwest Iran, and the study was conducted at the Faculty of Natural Sciences at the University of Tabriz. The study protocol was approved by the Ethics Committee of Tabriz University of Medical Sciences (IR.TBZMED.REC.1398.1064), and all participants provided written informed consent.
3.2. Genotype Analysis by PCR-RFLP
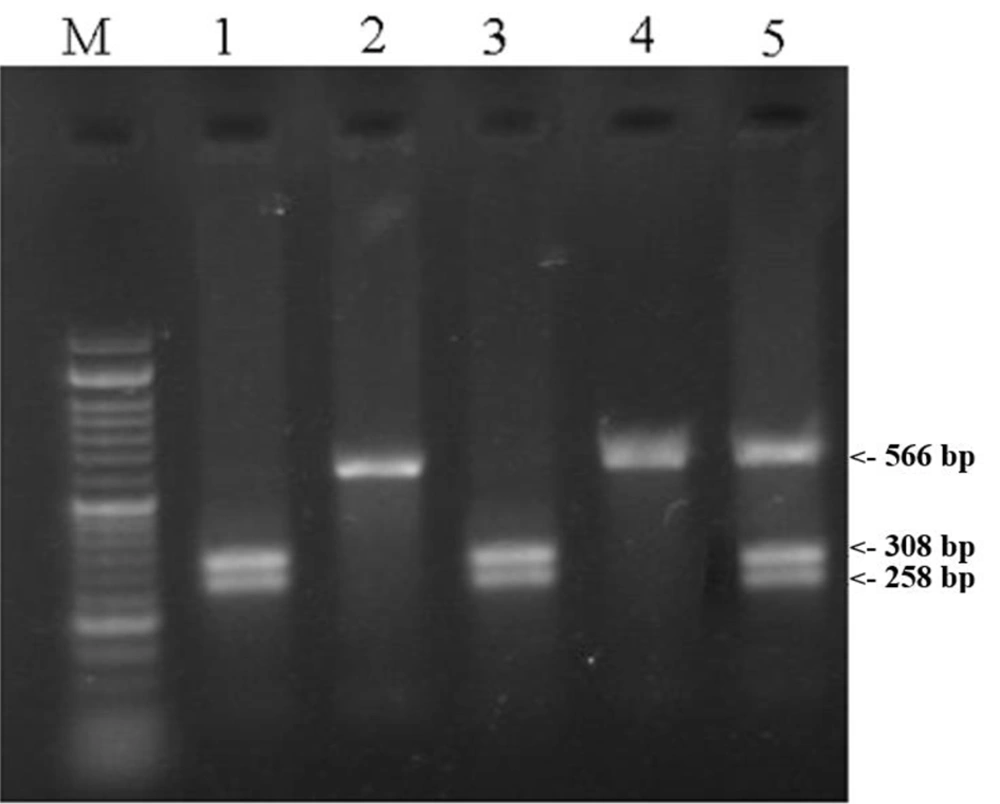
EDTA-treated peripheral blood samples were collected, and genomic DNA was extracted using magnetic nanoparticles from a commercial extraction kit produced by ZiAViZ (www.ziaviz.com). The polymorphic region was amplified using the forward primer 5´GGACAGCTCAGGGTAAGCAG3´ and the reverse primer 5´CCTGAGTACAGAGTTACGGG3´. Each PCR reaction was performed in a total volume of 15 µL, consisting of 1 µL of genomic DNA, 1 µL of each primer, 7.5 µL of PCR Master Mix (Ampliqon, Denmark), and 4.5 µL of ddH2O. The PCR was conducted on a thermocycler (Bio-Rad, USA) with the following program: An initial denaturation step at 95°C for 5 minutes, followed by 38 cycles of three steps (denaturation at 95°C for 30 seconds, annealing at 61°C for 30 seconds, and extension at 72°C for 30 seconds), and a final extension cycle at 72°C for 7 minutes. The PCR product was digested with 1.0 U of Pvu II restriction enzyme (Thermo Fisher Scientific, USA) for 1 hour and 45 minutes at 37°C and then electrophoresed on a 2% agarose gel. The polymorphic Pvu II site was detected by restriction fragment length polymorphism, producing fragments of 308 and 258 bp (TT homozygous genotype) or 566, 308, and 258 bp (CT heterozygous genotype). The 566 bp fragment (CC homozygous genotype) did not contain the site (Figure 1).
3.3. Statistical Analysis
Statistical analyses were performed using SPSS (version 20.0) to determine the frequencies of genotypes and alleles. The chi-square test was used to assess Hardy-Weinberg equilibrium. A 95% confidence interval was applied, and a P-value of <0.05 was considered significant for all statistical tests.
4. Results
In this study, the PD-1.5 C/T SNP was investigated in 52 SLE patients and 53 healthy controls. The genotype distribution for SLE patients (T/T 7.69%, T/C 34.62%, and C/C 57.69%) and the control group (T/T 16.98%, T/C 35.85%, and C/C 47.17%) showed that only the control group conformed to Hardy-Weinberg equilibrium (Table 1). Statistical analysis also revealed significant differences in genotype frequencies between the case and control groups (P = 0.030). Individuals with the CC and CT genotypes were approximately 2.707 (95% CI: 2.216 - 3.027, P = 0.0001) and 2.130 (95% CI: 1.481 - 2.956, P = 0.003) times more susceptible to SLE than those with the TT genotype (Tables 1 and 2).
Abbreviations: HWE, Hardy-Weinberg equilibrium; ns, non-significant.
a The Fisher's exact test P-value.
b The chi-square P-value.
c P < 0.05.
Abbreviations: OR, odds ratio; CI, confidence interval.
a The chi-square P-value.
b Reference.
c P < 0.05.
The allelic frequencies for the T and C alleles were 25% and 75% in the case group and 34.90% and 65.09% in the control group, respectively. A significant difference in allele frequency was also observed between the case and control groups (P = 0.019). Individuals with the C allele were 1.612 times more vulnerable to SLE than those with the T allele (95% CI: 1.015 - 2.117, P = 0.04) (Tables 1 and 2).
5. Discussion
It has been shown that the PD-1-PD-L pathway is essential for maintaining peripheral self-tolerance and preventing autoimmune diseases such as SLE (11). PD-1 and its two ligands, PD-L1 and PD-L2, provide inhibitory signals that regulate T cell activation, immunopathology, and tolerance (13).
Multiple studies have indicated that polymorphisms in the PDCD-1 gene are associated with an increased risk of SLE (14-18). In this case-control study, we investigated the PD-1.5 (+7785T/C) variant in patients with systemic lupus erythematosus and in healthy controls in northwestern Iran.
We found a significantly higher incidence of the PD1.5C/C and PD1.5C/T genotypes in SLE patients compared to healthy subjects, while the frequency of the PD1.5 T/T genotype was lower in SLE patients. These findings suggest that the PD1.5 T/T genotype may be protective against SLE, whereas the PD1.5 C/C and C/T genotypes may induce immune system down-regulation and predispose individuals in northwestern Iran to the disease. Additionally, we observed that the frequency of the PD1.5C allele was associated with SLE.
A meta-analysis study by Lee et al. (19) reported an association between the PD1.5C allele and SLE susceptibility among Europeans, but not Africans or Latin Americans. However, Gao et al. (20) found no association between the PD1.5 polymorphism and an increased risk of SLE in the Caucasian population. In another study, Abo El-Khair et al. (21) indicated that the prevalence of PD1.5C/T alleles was not significantly different between SLE patients and control subjects in Egyptian women. However, the PD1.5C/C and PD1.5C/T genotypes were significantly associated with SLE susceptibility. Additionally, they proposed that the PD1.5C/T polymorphism might only be in linkage disequilibrium with the actual causative allele, PD1.3G/A, or other unstudied locations, rather than being the causal allele itself. Chua et al. (22) also investigated this SNP in the three main ethnic groups of the Malaysian population and observed a significantly higher incidence of the PD1.5C/C genotype in Indian SLE patients and Malay controls. However, the PD1.5C/T genotype appeared to predispose Malays to SLE but had a protective effect in Indians (P < 0.01). Genetic variables implicated in the pathophysiology of SLE, as well as interactions with environmental factors, may account for variations in the correlation between polymorphisms and disease across different populations (22).
Although we reported an association between the PD1.5 polymorphism of the PDCD1 gene and SLE, this study has several limitations. The sample size was relatively small, consisting of 52 SLE patients and 53 healthy individuals, and further studies with larger populations are needed to confirm or refute our findings. Another limitation is that the study focused only on a single nucleotide polymorphism, even though there are multiple polymorphisms in SLE-related genes.
Despite these methodological limitations, this study also has several strengths. The control and SLE groups were similar in age, reducing potential confounding variables. To address the issue of population heterogeneity, which is a factor to consider in the Iranian population, sampling was conducted exclusively among individuals of Azeri descent.
Overall, we found that the PD1.5C/C and PD1.5C/T genotypes, as well as the PD1.5C allele of the PDCD1 gene, were significantly associated with systemic lupus erythematosus in the northwestern Iranian population.