1. Context
Influenza and coronavirus are two RNA viruses that have caused significant pandemics over the years (1, 2). Vaccination is undeniably the most effective approach to fighting viral infections. Although there are several available and approved vaccines against influenza and coronavirus, their protection may be temporary due to the rapid mutations of these viruses, and the risk of reinfection remains. Studies have shown that complementary herbal remedies play an important role in the prevention and treatment of diseases. Researchers are actively seeking to discover new low-cost and druggable bioactive molecules with antiviral properties and few side effects from natural sources. Additionally, knowledge of these viruses at the molecular level is rapidly expanding to identify druggable targets in novel subtypes of the viruses (3).
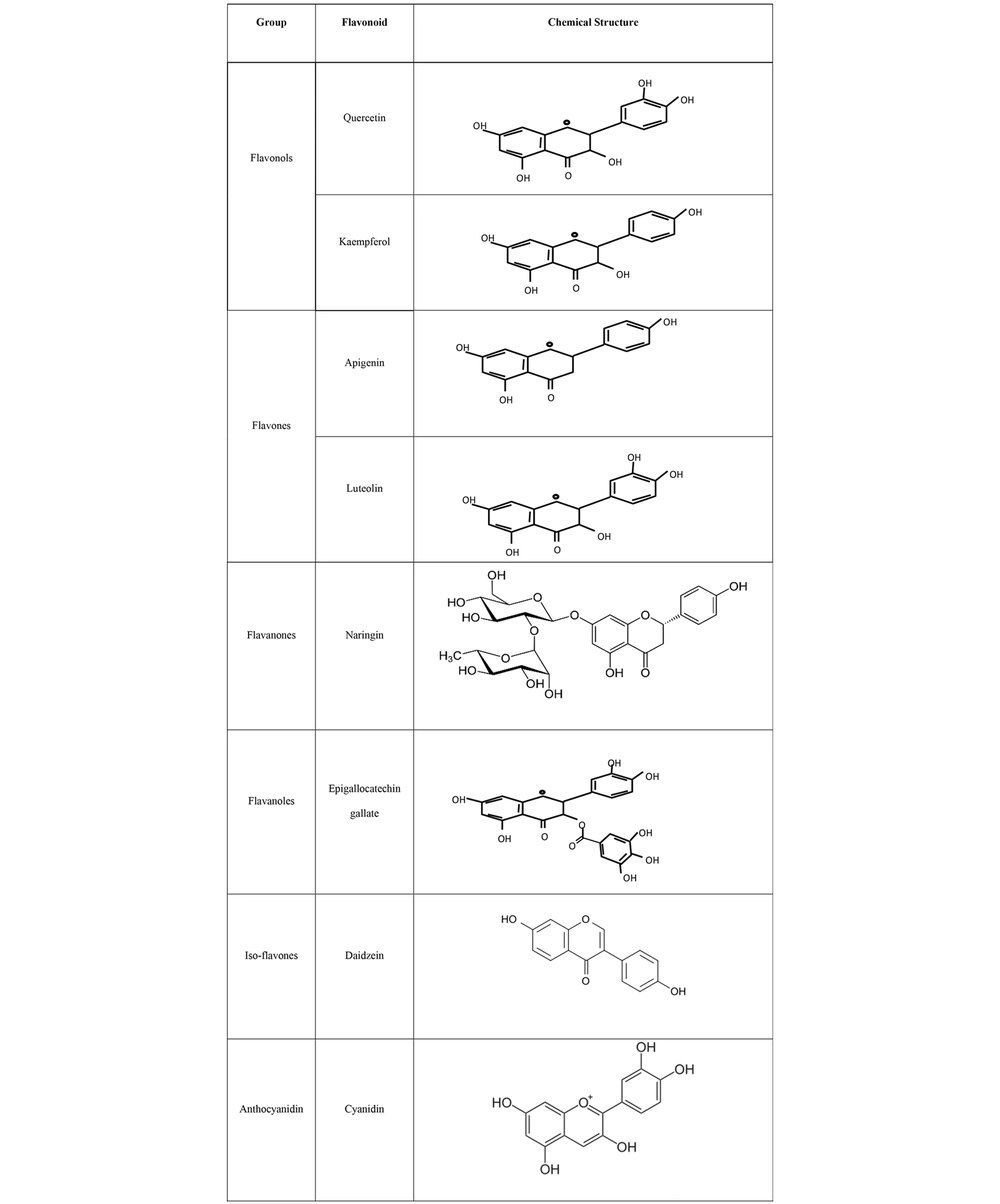
Flavonoids are a significant group of natural products belonging to a large class of plant secondary metabolites with polyphenolic structures, and they are mostly found in vegetables and fruits (4). The structure of flavonoids consists of a 15-carbon skeleton and two benzene rings bonded together by a heterocyclic ring containing oxygen. Based on their molecular and chemical structures, flavonoids are subdivided into six groups: Flavanones, flavones, flavonols, isoflavones, anthocyanidins, and flavanols (Figure 1) (5). A wide range of medicinal plants, such as lettuce, kale, onions, tomatoes, berries, apples, and grapes, are rich in flavonoids (4, 6). The main biological and health benefits of flavonoids include their antioxidant, anti-inflammatory, antibacterial, antifungal, and anticancer activities through various molecular mechanisms and pathways, such as increasing free radical scavenging by covalently binding to proteins and releasing one or two electrons (7).
Some important flavonoid compounds with antiviral activities include quercetin, kaempferol, apigenin, chrysin, naringin, luteolin, amentoflavone, epigallocatechin, epigallocatechin gallate, and gallocatechin gallate (8, 9), which mainly have antiviral activities (10). The antiviral activities of flavonoids are mainly related to their hydroxyl groups, which are able to stimulate the host cell defense system and inhibit virus binding and penetration (11).
Influenza and coronavirus undergo many genetic changes, resulting in various strains with different pathogenicity and transmission abilities. Recently, numerous computational and experimental studies have addressed the antiviral activity of flavonoid derivatives against influenza and coronavirus (10, 12). In this context, the present study reviews the pathogenesis of these two important viruses and the role of flavonoids in host-virus attachment and entry. It also exclusively summarizes the efficacy of flavonoids, their derivatives, and combination therapies targeting the enzymes and proteins of influenza and coronavirus.
2. Virus’s Entrance Mechanisms
Influenza and coronavirus infect host cells by specifically interacting with viral molecules and host receptors (13). To increase the efficacy of entry and ensure successful infection, viruses must overcome environmental obstacles, such as diffusion thermodynamics. Therefore, viral ligands can bind to host co-receptors through non-covalent and low-affinity electrostatic interactions. This attachment causes the two membranes to remain close to each other, allowing their surface entry receptors to interact more effectively. In the next step, viruses breach the cell’s phospholipid bilayer (14).
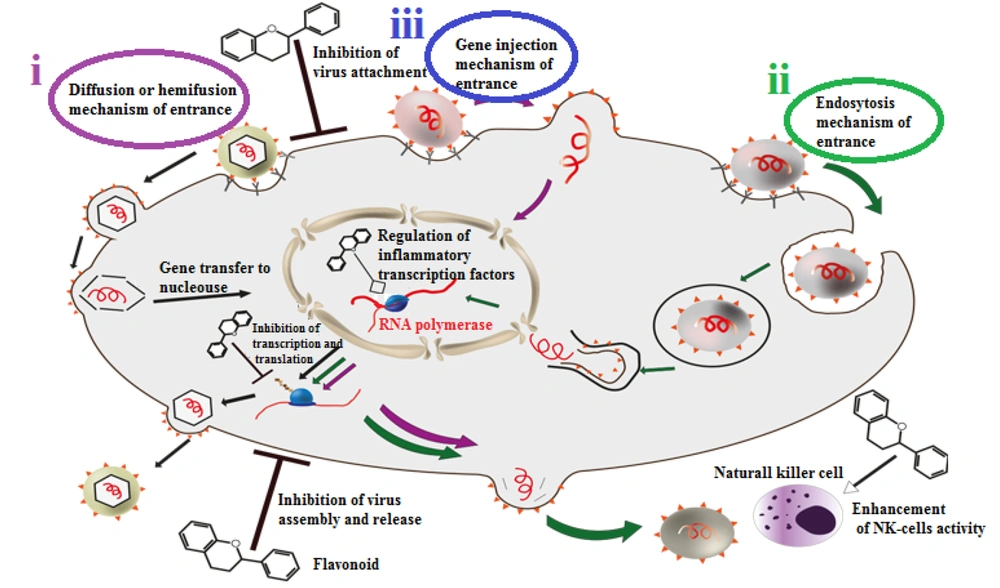
There are three main techniques for viral entry (Figure 2).
Three main mechanisms of viral entrance and some different antiviral effects of flavonoids. (i) diffusion or hemifusion: This method is special for enveloped viruses. They bind to their host cells via their ligand and receptors, virus envelope and host membrane would join and then the non-enveloped virus would release to the cell; (ii) endocytosis: In this method, host cell would recognize pathogenic viruses as harmless particles and swallow them like vesicles. Then viruses would reach the cytoplasm via endosome disengage process; (iii) gene injection: In this method, virus’s genome is sufficient for infection. The virus would stay on the host cell membrane and inject its genetic material.
i) Diffusion or hemi-fusion: This is a membrane interaction specific to enveloped viruses. Secondary receptors may mediate the connection between the cell membrane and the viral envelope. Then, the non-enveloped virus releases its content into the cell. This mechanism can be subdivided into low pH fusion, like influenza fusion, and pH-independent fusion, as seen in retroviruses (15).
ii) Endocytosis: This method is mainly specific to non-enveloped viruses, but some enveloped viruses can also use it. In this process, viruses disguise themselves as harmless particles, and host cells engulf them as vesicles. The virus then reaches the cytoplasm by disengaging from the endosome due to low pH and protease exposure.
iii) Gene injection: This technique is primarily used by viruses whose genome is sufficient for infection. In this method, the virus remains on the host cell membrane and injects its genetic material (15).
3. Influenza Viruses
Influenza, which causes seasonal epidemics worldwide, is an enveloped RNA virus with negative-sense genomic material. The influenza virus belongs to the Orthomyxoviridae family, which is categorized into four subgroups (A, B, C, and D). Subgroups A, B, and C are responsible for human infections. In infected cells, the influenza A virus can trigger the apoptosis pathway of receptor-interacting serine/threonine-protein kinase 3 (RIPK3) and mixed lineage kinase domain-like pseudokinase (MLKL)-dependent necroptosis (16). Differences in the antigenic properties of two viral surface proteins, hemagglutinin (HA) and neuraminidase (NA), divide the influenza A virus into subtypes H1 to H18 and N1 to N11 (17).
Currently, essential anti-influenza drugs include neuraminidase inhibitors (Zanamivir, Peramivir, and Oseltamivir), M2 and BM2 inhibitors (Rimantadine and Amantadine), and messenger RNA inhibitors (Ribavirin). Ribavirin acts as a nucleoside analog and can block inosine monophosphate dehydrogenase (IMPDH) (18, 19).
It has become increasingly clear that the first step for influenza infection is binding to and penetrating host cell receptors. Therefore, the external part of influenza virus HA molecules seeks host superficial sialic acid (SA) particles as their receptors. However, some low-molecular-weight molecules, such as annexin, can act as influenza receptors (17, 20).
Sialic acid is a 9-carbon monosaccharide with a negative electrical charge that can bind to glycoproteins and glycolipids through its terminal site. On average, nearly 60 genes encode different SA biosynthesis pathways, leading to various types of SAs. The variety in SAs is one factor that contributes to the different susceptibilities of host cells to influenza viruses (21, 22).
For example, the HA molecule of the human influenza virus has a greater affinity for SA-α-2,6-GA-terminated saccharides, influenced by its Leu 226 residue. SA-α-2,6-GA is more commonly expressed in the epithelial cells of the nasal passages, trachea, and bronchi. In contrast, the HA molecule of the avian influenza virus has a higher affinity for SA-α-2,3-GA-terminated saccharides, which is influenced by its Glutamine (Gln) 226 residue and is more highly expressed in the epithelial cells of the bronchioles and alveoli (23, 24).
These observations help explain the findings of van Riel et al., who discovered that the human influenza virus typically infects the upper portions of the respiratory tract, while the avian influenza virus infects the deeper parts of the respiratory tract (25).
Hemagglutinin, an influenza surface glycoprotein, binds to host cell SA molecules. The virus then enters the host cell through endocytosis and lysosomes. Hemagglutinin has two subunits: HA1 and HA2. The HA2 subunit is the stem part of the HA molecule and plays a significant role in the binding process, while HA1 is the molecule's head, which carries the receptor-binding domain with four different antigenic parts (26). Hemagglutinin is the part of the virus recognized and targeted by immune system antibodies, and it often undergoes many antigenic changes that can increase the severity of the disease or the virus's binding affinity (27).
The influenza virus enters the host cell via endocytosis. The low pH environment of the endosome causes HA molecules to change their conformation, leading to the fusion of the viral surface and the host cell's endosomal membrane. This fusion releases the viral genetic material into the cytoplasm, which is then transported to the nucleus. Subsequently, due to the hydrolysis process, the newly assembled virion is able to exit the host cell with the help of NA molecules. Neuraminidase is a transmembrane protein with catalytic activity for the terminal glycoproteins and glycolipids of SA in the host cell. Neuraminidase's sialidase activity disrupts the interactions between SA and HA molecules, which is essential for the release of the newly formed virion from the infected cell (28, 29). It has been reported that NA can also enhance cell-virus fusion (29).
Hemagglutinin, SA, and NA molecules are the main targets of flavonoid compounds for inhibiting the entry of the virus into the host cell. In the following sections, we will review and discuss information about flavonoids and their molecular interactions that prevent the attachment and entry processes.
4. Coronavirus
Coronavirus, from the Coronaviridae family, is a single-stranded, positive-sense RNA virus with an envelope containing a nucleocapsid. The RNA genome of the coronavirus includes essential genes such as replicases (Rep1a and Rep1b), spike (S), envelope (E), membrane (M), nucleoprotein (N), and additional genes coding for non-structural proteins (30). Its basic genome orientation is 5'-Rep1a-1b-S-E-M-N-3' (31). Different subclasses of coronavirus can cause severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and the new pandemic COVID-19. Additionally, zoonotic coronaviruses can infect a wide range of hosts, including avian species, swine, and humans. They are responsible for a range of respiratory problems, from mild symptoms like the common cold to more severe conditions like bronchiolitis and acute pneumonia (32).
Severe acute respiratory syndrome coronavirus (SARS-CoV-1) was responsible for the first coronavirus-caused epidemic in 2002/2003, with a mortality rate of 3 - 6% and up to 43 - 55% for older patients or those with underlying conditions (33, 34). The SARS virus life cycle begins with virion attachment to its specific membrane receptors, and through envelope and cell membrane fusion, its nucleocapsid enters the host cell. After replication and protein synthesis, new nucleocapsids form in the rough endoplasmic reticulum (RER). Mannose-rich carbohydrates are added to S proteins in the RER (35). Finally, new nucleocapsids gain their envelopes from smooth vesicles synthesized by the Golgi apparatus, completing the assembly of new virions (36).
The SARS-CoV-1 S protein recognizes the host's angiotensin-converting enzyme 2 (ACE2) receptors for attachment to its target cell, which can be a target for designing vaccines and inhibitor drugs. Angiotensin-converting enzyme 2 is a zinc metallopeptidase that plays a role in the renin-angiotensin system and has blood pressure regulatory activities. The SARS S molecule is a type I transmembrane protein composed of 1,255 amino acids (aa) and has four primary domains (35). S1 and S2 are large external domains that are essential for virus attachment and fusion. The other two smaller domains are the transmembrane and carboxyl-terminal cytoplasmic subdomains, which penetrate the viral envelope and anchor the S protein (37). Evidence suggests that the binding domain region for the S protein is located between aa-318 and aa-510, which has a high binding affinity for ACE2 receptors (38). Certain residues between aa-450 and aa-490 play crucial roles in optimal virus-host attachment. Other viral surface factors, including dendritic cell-specific C-type lectin (DC-SIGN) and related proteins of DC-SIGNR, may also affect S-ACE2 binding interactions and enhance the infection rate (35).
Middle East respiratory syndrome coronavirus (MERS-CoV) was responsible for the second major epidemic in 2012. In addition to respiratory tract diseases, MERS-CoV caused damage to other organs, such as the kidneys and liver, with a death rate of 34.9% (39). The MERS-CoV S protein recognizes and interacts with dipeptidyl peptidase 4 (DPP4), also known as the CD26 glycoprotein. This glycoprotein is a member of the type II transmembrane protein family, consisting of 766 amino acids, and is highly expressed in the prostate, liver, small intestine, and kidney's dimeric cells (40). Its extracellular domain contains an eight-bladed β-propeller at the N-terminal and an α/β hydrolase domain at the C-terminal. Blades 4 and 5 are the main regions involved in binding to the MERS-CoV receptor-binding domain (RBD) (41). Some host cell proteases, such as furin, cleave the spike protein into S1 and S2 subunits, a process necessary for binding and fusion (42). The S1 subunit consists of a core subdomain and a receptor-binding motif (RBM) that binds to DPP4 molecules, while the S2 subunit facilitates membrane fusion. During membrane fusion, the S2 subunit undergoes a conformational change, detaches from the S1 subunit, and exposes a fusion peptide with hydrophobic characteristics, ultimately allowing the two membranes to fuse (43). The S-RBDs of MERS and SARS coronaviruses are highly similar in core subdomain structure, but their amino acid sequences have low homology (41).
The newest member of the coronavirus family, SARS-CoV-2 (known as COVID-19), was first identified on 12 December 2019 and is characterized by mutations in its DNA sequence (44). Its human-to-human transmission ability is higher than that of other coronaviruses, with a mortality rate of approximately 2 - 4%. This new coronavirus can infect the same host cells as MERS and SARS and has a higher binding affinity for upper respiratory tract epithelial cells (45). The SARS-CoV-2 S protein has a trimeric structure that is cleaved into S1 and S2 subunits, with its amino acid sequence being about 76% identical to that of the SARS-CoV-1 S protein (46).
The S2 subunits of SARS-CoV-1 and SARS-CoV-2 have a 91% sequence alignment. However, there are notable differences between their S1 subunits and even more significant differences between the S1 subunits of MERS and SARS-CoV-2, which may explain COVID-19's broad binding potential for a variety of cell receptors, such as ACE2, CD26, Ezrin, glucose-regulated protein (GRP78), and cyclophilins (47, 48). Evidence suggests that the RBD of the SARS-CoV-2 S protein can strongly bind and interact with the human ACE2 receptor, which serves as the primary receptor for COVID-19 (46, 49-51). After attachment and membrane fusion, COVID-19 releases its genetic material into the host cytoplasm. The RNA genome then replicates and translates into essential and non-essential viral proteins, ultimately forming a new virus with the replicated RNA and synthesized proteins (52).
Since S proteins and their related receptors are ideal targets for drug design, many molecular docking studies have been conducted to examine the potential of natural compounds in inhibiting these molecules. Among these, flavonoid compounds have shown promising results (53-57).
5. Flavonoids Compounds
In recent decades, numerous studies have demonstrated that herbal compounds have positive effects on cardiovascular diseases, obesity, diabetes, and cancer, and they can reduce the mortality rates of these diseases. As a result, the consumption of herbal compounds is gaining significant attention worldwide. Flavonoids are among the beneficial herbal compounds that are abundant in plants. According to various studies, there is a direct link between consuming flavonoid-rich diets and improved life expectancy and human health (58).
Flavonoids are synthesized from phenylalanine and acetic acid through the shikimic acid pathway (59). The main structure of flavonoids consists of 15 carbons with two benzene rings. These rings are connected by a heterocyclic ring containing oxygen. As mentioned earlier, flavonoids are subdivided into six groups based on parameters such as molecular and chemical structures, the hydroxylation and glycosylation arrangement of the rings (5), and the degree of oxidation and unsaturation of the linking chains (9). These groups include flavanones, flavones, flavonols, isoflavones, anthocyanidins, and flavanols. The structural differences among these groups result in various biochemical properties, including anti-inflammatory, anticancer, antioxidant, anti-aging, and cardioprotective effects (4, 60, 61). Additionally, flavonoids have the ability to regulate the function of different cell enzymes (62). Their antioxidant properties arise from their structural hydroxyl groups and hydrogens (63). They can donate hydrogens to neutralize free radicals or prevent their formation (5). The anti-aging and cardioprotective properties of flavonoids are also related to these antioxidant activities (11).
The anti-inflammatory properties of flavonoids are associated with the deconjugation of their structural glycosides to aglycones (64). Flavonoids can inhibit various inflammatory factors such as phosphodiesterase 4 (PDE4) (65), nuclear factor kappa-B (NF-κB), pro-inflammatory transcription activator factor (AP-1) (66), and cyclooxygenase 1 and 2 (COX-1 and COX-2) (67). Flavonoids with anticancer properties can inhibit oxygen supply and some protein kinases, disrupt necessary nutrients (7), and interfere with the accumulation of vital growth and transcription factors and receptors (68). Additionally, they can interrupt angiogenesis and induce apoptosis by inhibiting the attachment of the 70 kilodalton heat shock protein (HSP70) promoter to heat shock factor 1 (HSF1) (69).
Studies have also shown that flavonoid compounds have many protective properties against human viral pathogens caused by both DNA and RNA viruses (10, 70). The mechanisms of flavonoids against viral infections include prophylactic inhibition and indirect inhibition through interactions with the immune system, blocking virus-host connections and entry by interacting with and attaching to different proteins and molecules, and inhibiting a series of intracellular actions such as virus replication, transcription and translation, assembly, packaging, and release from the host cell (71). Therefore, they can be valuable resources for drug design and complementary treatments alongside chemical drugs due to their numerous biological properties, abundance in the regular daily diet, and minimal side effects.
It is worth noting that all these biological benefits and safety profiles of flavonoids apply to their standard doses, but at high doses, they may be harmful. It has been reported that flavonoids can exhibit pro-oxidant activities instead of antioxidant effects, causing DNA damage, apoptosis, and cytotoxicity. They can also have adverse regulatory effects, as they may bind to different receptors and proteins' ATP-binding sites and interact with intracellular signaling pathways (72).
6. Flavonoids and Different Human Viruses
To better understand and expand on the subject of this article, we first briefly reviewed the effects of some bioflavonoid compounds on human pathogenic viruses. HIV is one of the most extensively studied viruses in this context. HIV infection begins with the binding of the virus envelope's gp120 subunit to T-cell surface CD4 molecules and CC chemokine receptor 4 (CCR4) or C-X-C chemokine receptor type 4 (CXCR4) co-receptors. Following this, the envelope gp41 subunit becomes active by forming a six-helix bundle conformation, facilitating the fusion of the virus and target cell. Previous analyses have shown that tannin (73) and theaflavin (TF), a flavonoid extracted from black tea (74), can inhibit the formation of the active gp41 form, thereby disrupting the fusion and entry of the virus into the host cell. These compounds do not affect gp120-CD4 binding or the activity of CXCR4 and CCR5 co-receptors. However, the flavonoid baicalin can block HIV-1 envelope-mediated fusion by interacting with CCR5 and CXCR4 co-receptors, which is believed to be its mechanism of action (75).
Epigallocatechin-3-gallate (EGCG) has polyphenolic rings that bind directly to the D1 domain of the CD4 molecule on the T-cell surface, inhibiting the attachment of gp120 molecules. Molecular modeling studies demonstrate that the amino acids Phe43, Arg59, and Trp62 of CD4, which are interaction sites for gp120, have an appropriate binding affinity to EGCG (76). Epigallocatechin-3-gallate is one of the most-studied flavonoid compounds in this context. For example, molecular docking and dynamic simulation studies have revealed that EGCG interacts with different molecules on the Zika virus surface, and these interactions block the membrane fusion process (77). The study by Sharma et al. supports the claim that the likely mechanism of EGCG in blocking virus entry is related to the membrane fusion process by interacting with various viral surface molecules (78). The galloyl moiety (ring D) of EGCG can also inhibit the entry of herpes simplex virus (HSV) through interactions with its essential envelope glycoproteins, gB and gD (79). In the case of hepatitis C virus (HCV), EGCG exhibits anti-infection effects with a possible mechanism of entry inhibition during the early stages of the binding process, rather than at the fusion stage (80). Similarly, two flavonoids, sorbifolin and pedalitin from Pterogyne nitens, have entry-blocking properties against HCV. Further studies have revealed that pedalitin can block virus entry by up to 80% through direct interactions with viral structures and up to 72.2% through interactions with host cells. In contrast, sorbifolin can interact with viral structures and block virus entry by up to 38.2% (81).
Further molecular docking-based analyses suggest that different bioflavonoids, including baicalin, fisetin, hesperidin, naringenin, naringin, quercetin, and rutin, can bind to the dengue virus's envelope proteins through hydrogenic, ionic, and hydrophobic interactions, thereby blocking the virus's entry into the host cell. Among all the mentioned bioflavonoids, quercetin had the highest affinity and the fewest side effects (82). Additionally, quercetin pentaacetate (an acetylated derivative of quercetin) shows virucidal activities against the human respiratory syncytial virus (RSV). In-silico analyses suggest that the mechanism of action of quercetin is its interaction with the virus’s F protein, which plays a critical role in virus fusion (83).
6.1. Flavonoids and Influenza Virus
As mentioned earlier, inhibiting the attachment of the influenza HA molecule to host SA receptors is the primary mechanism for preventing virus entry. Hemagglutinin inhibition assays and immunofluorescence staining have demonstrated that different derivatives of TF with 80% purity, including TF-3-G and TF-3'-G, can block the binding and entry of influenza viruses H1N1 and H3N2 to guinea pig erythrocytes. Their probable mechanisms include the direct binding of TF to HA, blocking the SA receptors, and indirectly inhibiting vRNP synthesis and its localization in the nucleus (84). Similarly, 3-2'-Dihydroxyflavone (3-2'-DHF) and 3-4'-Dihydroxyflavone have HA and NA inhibition properties in MDCK cells (85).
Using surface plasmon resonance (SPR) and microscale thermophoresis (MST) assays, quercetin, an aglycone flavonoid from the flavonol subgroup (86), has shown a binding affinity for HA molecules. Further studies and assays revealed that quercetin can bind to the HA2 subunit of the HA molecule and inhibit virus entry. It also reduces the transcription of viral HA mRNA in host cells (87). Two elderberry flavonoids, 5,7-dihydroxy-4-oxo-2-(3,4,5-trihydroxyphenyl) chroman-3-yl-3,4,5-trihydroxycyclohexanecarboxylate and 5,7,3',4'-tetra-O-methyl quercetin, can block virus-host attachment. Based on these compounds, 5,7,3',4'-tetra-O-methyl quercetin and racemic dihydromyricetin were synthesized, and all these compounds were found to bind to the viral envelope, particularly to the recognition and binding domains of the HA molecule, thereby blocking H1N1 virion entry into cells (88).
TSL-1, an extract from Toona sinensis leaves (TSLs), is a traditional herb in China and Taiwan. It contains several bioactive compounds, such as gallic acid (89), different derivatives of quercetin, kaempferol, and rutin (90), along with many other flavonoids and polyphenolic compounds. This traditional herb can impede H1N1 virus infection primarily by inhibiting attachment rather than viral penetration in MDCK cells. TSL-1 can also inhibit the transcription of adhesion molecule genes in A549 host cells (91). Alpinia katsumadai is another Chinese herbal medicine rich in diarylheptanoids, monoterpenes, sesquiterpenoids, chalcones, and flavonoids. It has a binding affinity for the surface HA of the H1N1 virus and can prevent the attachment of the virus to the host cell (92).
Cistus incanus extract from CYSTUS052, a Mediterranean plant rich in polyphenol compounds, has been shown to inhibit HA activities in pre-treated influenza viruses (93). Oligonol, a polyphenol extract from lychee fruit, contains several main flavonoids, including catechin and proanthocyanidin. This extract also exhibits anti-influenza activity by directly binding to and inhibiting the HA molecule (94). Additionally, molecular docking studies have shown that curcumin and its derivatives, such as desmethoxycurcumin and bisdemethoxycurcumin, have a binding affinity for the HA molecule with sufficient binding free energy (95).
Disrupting the virus's membrane and envelope and altering its morphology is another method to block viral entry. In this context, some polyphenolic extracts have shown beneficial effects. According to observations by Kim et al., EGCG can alter the morphology and size of influenza viruses A and B and block their penetration before virus-cell fusion. Epigallocatechin-3-gallate can induce physical damage, especially to the phospholipid bilayer, causing the virus to lose its rigidity and ability to move forward (96).
Pomegranate (Punica granatum, Punicaceae), a native fruit of Iran, Afghanistan, India, and Mediterranean countries, is rich in phenolic and anthocyanin compounds and exhibits powerful antioxidant and antiviral activities. Its key phenolic compounds include punicalagin, caffeic acid, ellagic acid, and luteolin. Transmission electron microscopy (TEM) analyses have shown that pomegranate polyphenols lead to abnormal and broken enveloped glycoprotein particles in the H3N2 influenza virus. In contrast, for H1N1 particles, both fragmented particles and unaffected regions with standard enveloped glycoproteins have been observed (97).
H9N5 viruses pre-treated with hydroxytyrosol (HT), an abundant phenolic compound in olive leaves and fruits, showed morphological changes and disrupted envelope membranes in TEM analyses. Treated H9N5 viruses lacked a surface spike layer, and instead of confined HA molecule localization, they showed a more dispersed form (98).
Hemagglutination is a mechanism that results from the interactions of some enveloped virus surface glycoproteins with red blood cells. EGCG and epicatechin-gallate (ECG), but not epigallocatechin (EGC), can inhibit the hemagglutination ability of the influenza virus (99). In chicken red blood cells infected with the influenza A virus, punicalagin also showed strong cell-growth and agglutination inhibition properties (100).
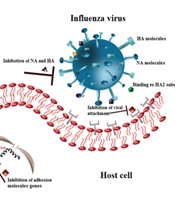
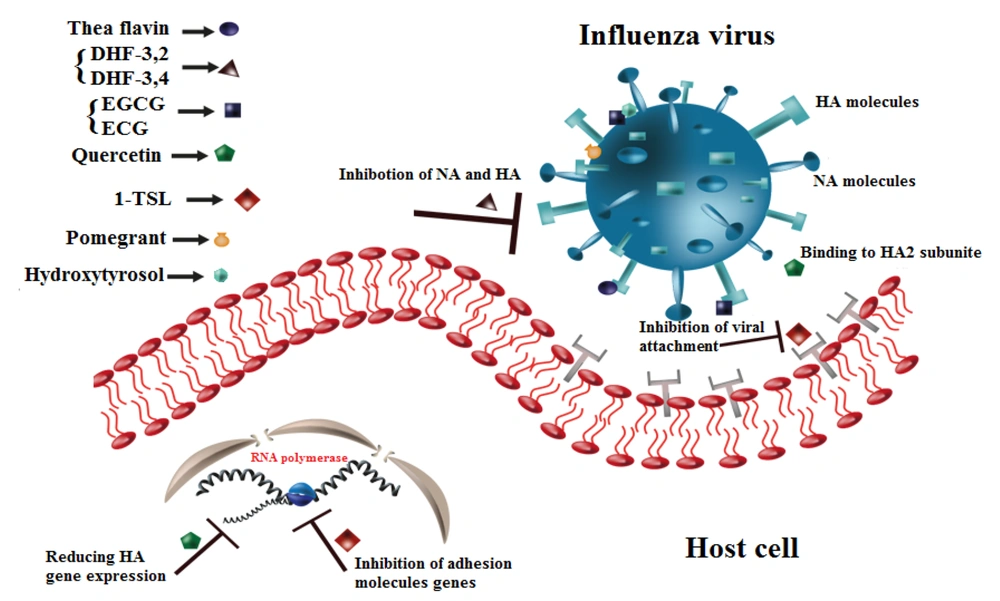
Table 1 and Figure 2 show the inhibitory properties of different flavonoids on various strains of the influenza virus. As illustrated in Figure 3, flavonoid compounds and their derivatives primarily have inhibitory effects on influenza molecules.
| Flavonoid | Influenza Strain | Strategy Model | Mechanism of Inhibition | Reference |
|---|---|---|---|---|
| Theaflavin with 80% purity; TF-3-G; TF-3'-G | H1N1; H3N1 | In-vitro | Binding affinity to HA or SA receptors, indirectly inhibition of synthesis and nucleus localization of vRNP | (84) |
| 3,2'-DHF; 3,4'-DHF | H1N1 | In-vitro + in-vivo in mouse models | Inhibition properties against HA and NA | (85) |
| Quercetin | H1N1; H3N2 | In-vitro | Binding to HA2 subunit of HA molecule, reducing the expression of HA mRNA | (87) |
| 3, 4, 5-Trihydroxyphenyl and 5, 7, 30, 40-Tetra-O-methylquercetin | H1N1 | In-vitro | Blocking the attachment of virus to the host cell and binding to viral envelope and inhibition of binding and entrance | (88) |
| TSL-1 | H1N1 | In-vitro | Inhibition of viral attachment and exprresion of adhesion molecule's genes | (91) |
| Alpinia, Katsamadia | H1N1; H9N2 | In-vitro | Binding to HA molecule and inhibition of attachment | (92) |
| CYSTUS-052 | Different avian and human influenza viruses | In-vitro + molecular basis analyses | Polyphenols binding to the viral surface and blocking the interactions between HA and receptors | (93) |
| Oligonol | H3N2 | In-vitro | Inhibition of HA attachment to the host cell via directly binding to it | (94) |
| Curcumin and its derivates | H1N1; H2N2; H3N2; H5N1 | Molecular docking + molecular dynamic simulation | Binding affinity for HA molecule with acceptable free energy | (95) |
| EGCG | Type A and B influenza virus | In-vitro | Inhibition of penetration via induction of phospholipid bilayer physical damages and morphological changes | (96) |
| Pomegranate | H1N1; H3N1 | In-vitro | Entry blocking via induction of morphological abnormalities | (97) |
| Hydroxytyrosol | H9N5 | In-vitro | Induction of envelope disruption and changing the surface spike layer aggregation model | (98) |
| EGCG; ECG | H1N1; H3N1; Type B influenza virus | In-vitro | Hemagglutination inhibition | (99) |
| Punicalugin | H3N2 | In-vitro | Inhibition of agglutination and growth of chicken RBC's infected cells | (100) |
The Blocking Properties and Mechanisms of Different Flavonoids on Different Strains of Influenza Virus
Entry blocking properties of different flavonoids on influenza virus. Theaflavin binds to Hemagglutinin (HA) or sialic acid (SA) molecules. DHF-'3,4 and DHF-'3,2 inhibit neuraminidase (NA) and HA molecules. Epigallocatechin-3-gallate (EGCG) and epicatechin-gallate (ECG) interact with HA molecule and change viral membrane physical activities, Quercetin binds to HA2 subunit of HA molecule and reduces the HA gene expression. TSL-1 inhibits the expression of adhesion molecule’s gene. Pomegranate and Hydroxytyrosol induce morphological abnormalities.
6.2. Flavonoids and Coronavirus Family
Based on in-silico and docking studies, which are among the most reliable analyses, some flavonoids extracted from citrus fruits show binding affinity for ACE2 molecules. Among them, naringin, with a docking energy of -6.85 kcal/mol and potential binding sites at TYR-515, GLU-402, GLU-398, and ASN-394, and hesperidin, with a docking energy of -6.09 kcal/mol and binding sites at LYS-562, GLU-564, and GLY-205, exhibited the highest binding affinity to the ACE2 enzyme. Naringin can alleviate the cytokine storm that usually occurs in severe cases of COVID-19 by inhibiting the expression of pro-inflammatory cytokines, including IL-1β, IL-6, COX-2, nitric oxide synthase, and high mobility group box 1 (HMGB-1) (53).
On the other hand, another study demonstrated that hesperidin and naringin did not have a desirable binding relationship with ACE2 receptors, but they did show desirable binding energy with the spike protein's RBD (54). Hesperidin, along with other flavonoids such as cannabinoids, rhoifolin, pectolinarin, morin, epigallocatechin gallate, and herbacetin, may be the best candidates for spike inhibition, as they exhibited the highest binding affinity for the spike protein (55, 56). Therefore, the RBD-S, PD-ACE2, and SARS-CoV-2 protease with a strong binding affinity for hesperidin are its important targets (57).
Quercetin is one of the most-studied flavonoids and has shown excellent docking value (-144.09 kcal/mol) for interaction with ACE2 receptors by forming a salt bridge with the LYS353 residue that interacts with the ACE2 ASP 38 residue (54). Quercetin, along with fisetin, also showed acceptable binding energy for the S2 subunit of the spike glycoprotein in docking experiments conducted by Preeti Pandey. Further investigations also revealed that these two flavonoids could bind to the ACE2-S complex and disrupt their interaction (101). Compounds such as curcumin, galangin, scutellarein, morin, silibinin, abacavir, myricetin, and epigallocatechin have also been considered spike inhibitors based on in-silico studies, and among them, baicalin is the most potent candidate. Epigallocatechin-3-gallate can also bind to ACE2 molecules (102).
Actenoside, an extract from Phlomis aurea, a native plant in Egypt with polyphenolic properties, has shown the highest binding affinity for the C-terminal domain of the spike glycoprotein's S1 subunit. It is important to note that other flavonoid compounds, such as luteolin and liriodendrin, showed binding affinity for both the human ACE2 receptor and the spike glycoprotein (103). Punicalin and punicalagin, which are the main polyphenolic extracts of pomegranate peel, have high binding affinity for both S and ACE2 molecules, making them likely candidates as COVID-19 entry blockers (104). Various flavonoids and polyphenolic extracts of propolis, which is produced by honeybees, also showed binding affinity for the S1 subunit of the spike protein with different docking scores. Among them, rutin, caffeic acid phenethyl ester, and pinobanksin exhibited the highest affinity (105).
To better study the effectiveness of flavonoids and polyphenolic compounds against SARS-CoV-2 virus entry and to confirm computational studies, many in-vitro investigations have been conducted. Recently, scientists treated some engineered HIV-Luc/SARS pseudo-typed viruses expressing the S protein with 130 small compounds. Among those compounds, luteolin and tetra-O-galloyl-β-D-glucose (TGG) successfully inhibited the S protein, with IC50 values of 2.86 and 9.02 μM, respectively. Luteolin analogs, such as quercetin, which have some structural similarities, also showed inhibitory effects against HIV-Luc/SARS pseudo-typed viruses with an IC50 of 83.4 μM (106).
According to an in-vitro study, baicalin has inhibitory properties against renin and ACE2, with IC50 values of 120.36 μM and 2.24 mM, respectively. Therefore, it appears that baicalin could inhibit the entry of these viruses by interacting with the ACE2 enzyme (107). Phenol-rich compounds, such as extracts from green tea and spirulina, were found to reduce the entry and infection of Vero-E6 cells with pseudo-typed viruses expressing the S protein of SARS, COVID-19, and MERS. When HEK293T cells expressing ACE2, DPP4, and the S1 subunit of the S protein were incubated with the same extracts, it was suggested that green tea could bind to and inhibit the S1 subdomain, although the inhibition mechanism of spirulina remains unknown (108).
Recently, it has been reported that Gene-Eden-VIR/Novirin has advantages in fighting against beta coronaviruses, especially SARS coronavirus and COVID-19. Its main ingredients include quercetin, cinnamon, liquorice, and selenium. Its functional mechanisms involve inhibiting cell entry and infection, replication, protease activity, and enhancing the immune system (109). With the development of new variants of SARS-CoV-2, Tran et al. examined the effects of phenolic-rich dandelion (Taraxacum officinale), a member of the Asteraceae plant family. Results from in-vitro experiments showed that dandelion could potentially inhibit the formation of different variants of the spike glycoprotein and ACE2 complex (110).
The transmembrane protease serine 2 (TMPRSS2) is a surface serine protease on the host cell that causes irreversible proteolytic cleavage of the spike protein, facilitating the viral-host fusion process (111). Docking analyses revealed that some flavonoids, such as neohesperidin, myricitrin, and quercitrin, could successfully bind and interact with the TMPRSS2 protein, leading to viral entry inhibition (112). Lutonarin, a natural compound rich in flavonoids, binds to three active sites of the TMPRSS2 molecule, including HIS-296, ASP-345, and SER-441, with a binding affinity of -8.8 (113). Additionally, the aforementioned extracts of punicalagin and punicalin can bind to the TMPRSS2 molecule and inhibit its activity (114).
Table 2 and Figure 4 show the blocking properties of different flavonoids on various strains of coronavirus. Interestingly, flavonoid compounds and their derivatives, unlike with influenza, primarily have an inhibitory effect on host cell molecules.
| Flavonoid | Target Molecule | Study Model | Mechanism of Action | Reference |
|---|---|---|---|---|
| Naringin, hesperidin | ACE2 | Molecular docking | Binding to TYR-515, GLU-402, GLU-398, and ASN394 protein binding site, Binding to LYS-562, GLU-564 and GLY-205 protein binding site | (53) |
| Hesperidin, cannabinoids, rhoifolin, pectolinarin, morine, epigallocatechin gallate, herbacetin | COVID-19 spike protein | Molecular docking | High binding affinity for spike protein | (55, 56) |
| Quercetin | COVID-19 ACE2 receptors | Molecular docking + pharmacological analyses | Forming a salt bridge with ACE2 receptors | (54) |
| Quercetin, fistein | Spike S2 subunit; ACE2/S complex | Molecular docking | Binding and interaction with spike molecule and ACE2-S complex | (101) |
| Baicalin; curcumin galangi Scutellarein | Spike protein | Molecular docking | Inhibition of spike protein | (102) |
| Actenoside; luteolin, liriodendrin | Spike S1 subunit; ACE2/ spike protein | Molecular docking | binding to C-terminal domain of spike S1 subunit | (103) |
| Punicalin, punicalagin | Spike molecule ACE2; TMPRSS2 | Molecular docking | Binding and interaction with molecules | (104) |
| Propolis extracts | S1: Subunit of spike protein | Molecular docking | Forming H-bond with spike protein | (105) |
| Luteolin,TGG | SARS spike protein | In-vitro | Inhibition of spike protein | (106) |
| Baicalin | Renin molecule | In-vitro | Inhibition properties for renin activities | (107) |
| Green tea, spirulina extracts | Spike protein | In-vitro | Binding to S1 sub domain of spike protein | (108) |
| Dandelion | Different variants of spike glycoprotein/ACE2 complex | In-vitro | Inhibition the formation of S/ACE2 complex | (110) |
| Neohesperidin, myricitrin, quercitrin | TMPRSS2 | Molecular docking | Binding and interactions with TMPRSS2 molecule | (112) |
| Lutonarin | TMPRSS2 | In-silico | Binding to active sites of the molecule | (113) |
The Blocking Properties and Mechanisms of Different Flavonoids on Different Strains of Coronavirus
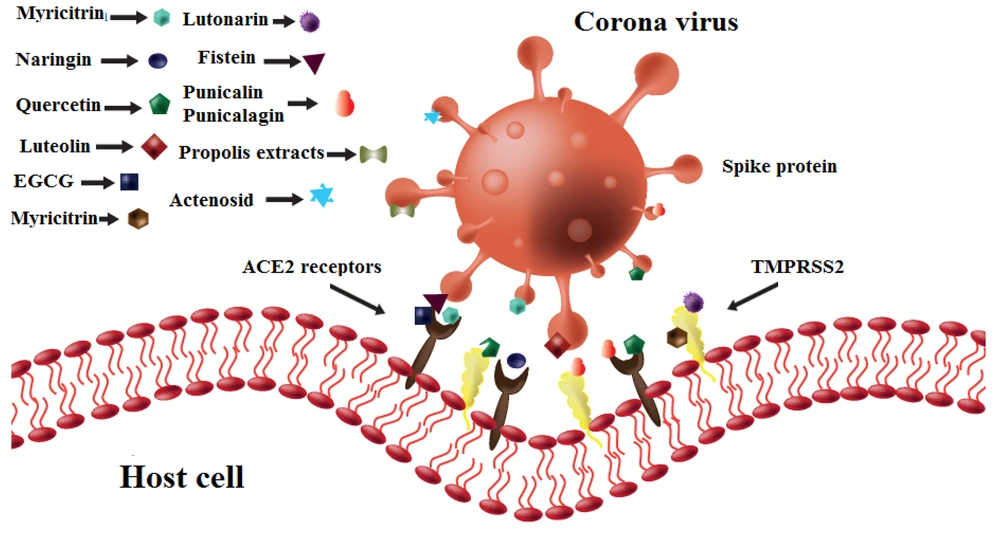
Entry blocking properties of different flavonoids on different strains of coronavirus. Hesperidin binds to spike proteins and ACE2 receptors. Naringin, quercetin, and epigallocatechin-3-gallate (EGCG) bind to ACE2 receptors. Quercetin, myricitrin, and lutonarin bind to TMPRSS2 molecules. Luteolin inhibits spike proteins.
7. Combination Therapy of Flavonoids and Antiviral Drugs
Despite the antiviral properties of flavonoids against influenza and coronavirus, they may be harmful and have side effects at high doses. To address this issue, many studies have suggested combination therapy. Using two or more drugs that target several viral proteins in combination treatments might offer various advantages, such as cost-effectiveness, greater impact and intensity, lower dosages, and a reduced risk of respiratory complications (19).
According to an in-vivo experiment, combination therapy of H1N1-infected mice with baicalein and ribavirin led to fewer inflammatory responses and less pathogenicity than when these drugs were administered alone, although the precise underlying mechanism was not identified (19). Baicalein and biochanin A can also amplify the anti-neuraminidase effects of zanamivir in H5N1-infected A549 cells (114). Iso-quercetin, a derivative of quercetin with glucose, and amantadine have synergistic effects in infected MDCK cells. An increase in the dosage of iso-quercetin resulted in a decrease in synergistic effects. Combination therapy with iso-quercetin and either amantadine or oseltamivir also inhibited drug resistance in infected cells and significantly decreased virus titer for up to 10 passages (115). A polyphenol extract from Geranium sanguineum can regulate the activity of host lung-secreted proteases and anti-proteases during influenza infection. Combining this extract with ε-aminocaproic acid (ACA), a protease inhibitor, created a synergistic effect and improved the therapeutic properties of ACA in the lung cells of mice infected with the influenza A virus (116). Furthermore, pomegranate, due to its anthocyanin flavonoids and other phenolic compounds, can enhance the ability of oseltamivir to reduce viral release from infected MDCK cells (100).
Several chemical drugs and agents, including chloroquine, hydroxychloroquine, lopinavir, ritonavir, ribavirin, corticosteroids, immunoglobulin therapy, and some immune-regulatory agents, could be candidates for coronavirus treatment (117). Recent evidence on the beneficial effects of remdesivir, an RNA-dependent RNA polymerase inhibitor, suggests that it could be the best choice for severe cases of COVID-19 (118, 119). A molecular docking study has evaluated the combination therapy of darunavir (an anti-HIV drug) and quercetin–3-rhamnoside. The results demonstrated that the main protease of COVID-19 was effectively targeted, and the total docking score was -14.83 (120). Additionally, two recent in-vitro and RCT studies have reported that ivermectin can inhibit COVID-19 replication and improve clinical parameters in patients. Other studies have suggested further benefits from the synergistic prophylactic effects of quercetin with ivermectin (121-123).
8. Conclusions
To treat influenza and coronavirus more effectively, we should move toward low-cost therapeutic approaches with fewer side effects. On the one hand, the recent increase in knowledge about the antiviral effects of natural compounds such as flavonoids, and on the other hand, the genetic changes that occur every year leading to the emergence of different strains of influenza and coronavirus with varying pathogenicity, have prompted us to collect recent studies in this field and take a closer look at the effects of flavonoids on these viruses.
As reviewed, the results of in-silico, in-vitro, and in-vivo studies have shown that flavonoid derivatives can play various roles in modulating different stages of viral infection, particularly in the attachment, fusion, and penetration stages, thereby suppressing the infection in its early steps. In addition, flavonoids can directly bind to and coat surface proteins and receptors on viruses and host cells or indirectly disrupt membrane integrity or reduce the transcription of related genes. Given the severe and life-threatening respiratory complications associated with both viruses, it appears that developing a new compound as a potent herbal-based antiviral treatment, adjunct, or complementary remedy with minimal adverse effects and the ability to simultaneously inhibit both viruses are within reach. However, the lack of sufficient information about COVID-19 is one of the limitations of our study, which requires further research.