1. Context
Coronavirus (CoV) is a respiratory-related infectious disease that has a single-stranded RNA genome of four groups: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. It is generally thought that mammals host Alphacoronavirus and Betacoronavirus while birds host Gammacoronavir. The virus can enter human cells via viral particles. These viruses bind to cellular receptors by S virus-mediated proteins, leading to the fusion of the virus into the cell membranes. Porcine delta coronaviruses (PDCoVs) belong to the family of delta coronavirus and the family Coronaviridae. Potential PDCOV interstitial transmission may occur between small wild mammals with pigs as well as between domestic pigs and birds with birds. Coronaviruses can cause diseases in birds and mammal species, including humans. It causes severe diarrhea, vomiting, and dehydration in infants (1-4). Apoptosis in PDCoV-infected intestinal epithelial cell membranes does not cause vacuole degradation and possible death, but necrosis is likely due to the cytolytic activity of the virus (5). A type of gamma coronavirus is infectious bronchitis virus (IBV) in birds that causes respiratory, reproductive, renal, and gastrointestinal disorders of nephritis. This virus first spreads to the trachea and then spreads to the internal organs (6, 7). The first coronavirus viruses were identified in 1983 with strains isolated from birds with the respiratory syndrome called the quail CoV (8, 9). Infectious bronchitis virus in the kidney results in cytopathological changes in the tubular epithelium associated with destruction, necrosis, and inflammatory response of the interstitial tissue (10). The bovine coronavirus (BCoV) is a beta coronavirus, is structurally similar to eukaryotic messenger RNAs (11). A coronavirus is widespread in 45% - 80% of wild and domestic cat species of all ages worldwide. This virus was detected in cats by experimental tests (12). Severe acute respiratory syndrome (SARS) appeared in humans in late 2002 in Guangdong, China, subsequently spread early the following year. Shortly after the virus spread to China, the causative agent quickly became known as a previously unknown type of coronavirus (13). In addition, it is thought that bovine coronavirus (BCV) may have jumped to human hosts by recombination with the influenza C virus, producing human coronavirus-OC43 (HCOV-OC43) (14). Many types of coronaviruses, including HCOV-229E, SARS-CoV, HCOV-OC43, and HCOV-NL63, can induce human infection. Studies of respiratory pathogenesis have been performed in this type of coronavirus (HCOV-229E). HCOV-229E infection in humans can lead to headaches, coughs, fever, and dysfunction of the nasal epithelium. HCOV-229E can enter the human cells through the N-aminopeptidase (hAPN) of the surface receptor (15, 16). The only notable exception in coronaviruses is the HCOV-NL63 variant that uses an angiotensin-2-converting enzyme (ACE2) to enter the cell (17). Most coronaviruses cause intestinal or respiratory infections, but some spread systematically. For example, the virus that induces mouse hepatitis (MHV) also infects other organs, including the liver and brain (18). Equine coronavirus (ECOV) was similar to hepatitis C-inducing coronavirus viruses. ECOV, through a caspase-dependent pathway, interacts with virus proteins, cell surface signaling factors and participates in both apoptotic pathways leading to cell death and mitochondrial cell death, causing apoptosis (19, 20). Feline infectious peritonitis (FIP) emerges when its host mutates occur that leads to immunodeficiency and uncontrollable infection (21, 22).
Acute kidney injury (AKI) is common in 25% of COVID-19 patients. The mortality rates are high in these patients. Some of the clinical studies indicate that the kidney is the store of SARS-CoV-2. The presence and activity of this type of coronavirus in this organ lead to proteinuria and hematuria. The pathophysiology mechanism of COVID-19 in inducing AKI could be directly associated with cellular injury by viral entry through the ACE2 receptor, which is highly expressed in the kidney during COVID-19. The direct impact of kidney damage caused by SARS-CoV-2 can be cited as an imbalanced renin-angiotensin-aldosterone system, pro-inflammatory cytokines, and thrombotic events. Moreover, other indirect mechanisms from SARS-CoV-2 that cause AKI during COVID-19 are hemodynamic alterations, right heart failure, hypovolemia, and nosocomial sepsis (23-25).
Therefore, according to the before researches and the pathological effects of coronaviruses on different organs of the body, this scoping review investigated the effect of these viruses, especially SARS-CoV-2, in inducing kidney damage and its probable cellular mechanism of action other than ACE2 receptor.
2. Evidence Acquisition
2.1. Present Study
This article is a scoping review based on Arksey and Omally’s method (26, 27). Present research may be useful for systematic review and meta-analysis studies. The PRISMA-SCR checklist designed by Tricco et al. (28) was applied to write this article. The present scoping review had six steps, as follows (27):
2.2. Identifying the Study Problem and Associate Article
To achieve the aim of the study, English articles in the PubMed database were searched until October 2020 by utilization of the MeSH Term system, including the four groups of search terms of coronavirus, COVID-19, and kidney and in combination with each other. The inclusion criteria were publication characteristics, language, and article type, such as primary and secondary, that revealed the correlation between coronavirus and kidney.
2.3. The Article Choice
The screening was done by reviewing the title and abstract of articles after transferring them to EndNote (X7) software. Some of the selected articles that did not comply with the aim of the study were removed. According to the PRISMA flowchart, two researchers investigated the full text of the articles in this phase of the study.
2.4. Data Expansion
The data charting was done and appeared in Appendix 1 (see Supplementary File) as a data extraction form.
2.5. Collecting, Briefing, and Showing the Results
The results were assessed based on the data extraction form. In this study, different types of coronavirus and its effect on kidneys were reported. Finally, the data were presented in tables and graphs.
3. Results
3.1. The Characteristics of Administrated Articles
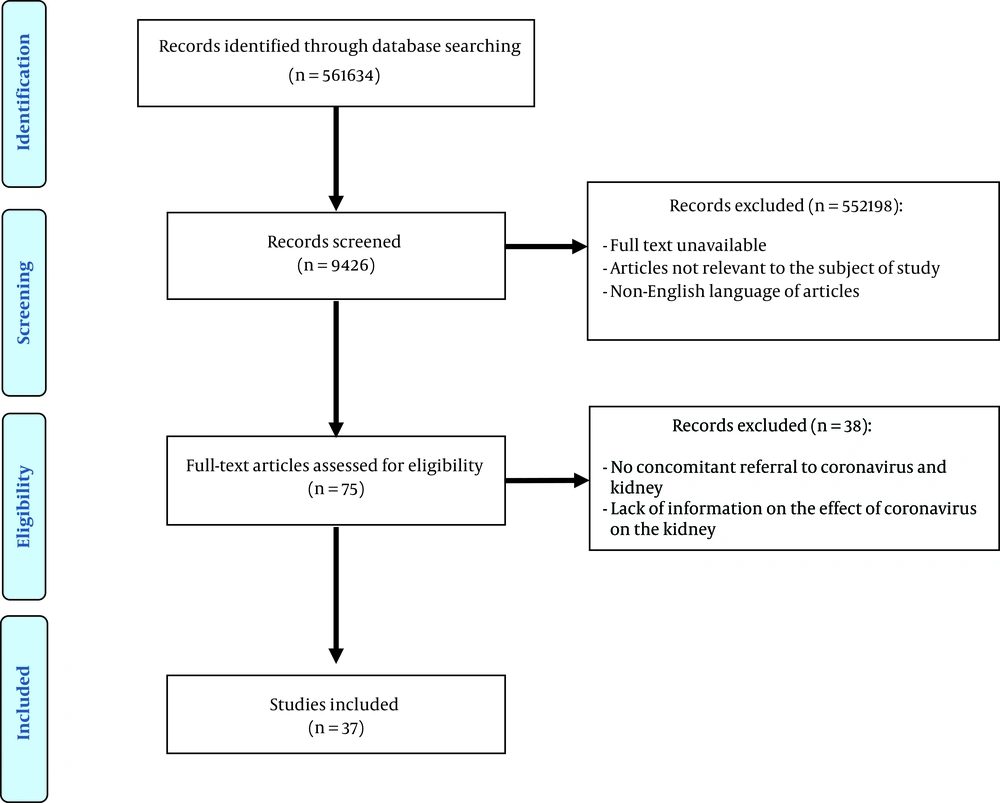
After deleting the similarity of articles (n = 561634), 9,426 of them were selected for the screening and title and abstract review. A total of 75 studies were selected, and 37 were remained (Figure 1). The properties of these articles were categorized and summarized in Table 1. In this study, 94% were paper studies, as well as 63% and 37% were primary and secondary researches, respectively. All of the manuscripts had been published in English, 54% of which were between “2010 - 2020” and 46% between “1990 - 2010”, and the study population was 24% human, 31% animal, and 45% cellular. Moreover, 8% of the study participants included studies on coronavirus, and 92% of studies included the effect of the virus on kidney tissue or cells. Also, the sample size in 31% of the articles was less than 50, 16% between 51 and 150, and 3% more than 150. The lowest sample size was 1, and the highest sample size was 284. Also, 3% of the studies on humans was less than 1-year-old, 3% of the studies on humans was more than 1-year-old, 23% of the studies on animals was less than 1-month-old, 5% of the studies on animals was more than 1-month-old, and 66% of the studies were not cited because of their cellularity and age.
| Number | Search Syntax | Results |
|---|---|---|
| 1 | (Coronaviruses[tiab] OR Deltacoronavirus[tiab] OR Deltacoronaviruses[tiab] OR “Munia coronavirus HKU13”[tiab] OR “Coronavirus HKU15”[tiab] OR (Coronavirus[tiab] AND Rabbit[tiab]) OR “Rabbit Coronavirus”[tiab] OR (Coronaviruses[tiab] AND Rabbit[tiab]) OR “Rabbit Coronaviruses”[tiab] OR “Bulbul coronavirus HKU11”[tiab] OR “Thrush coronavirus HKU12”[tiab]) | 4,675 |
| 2 | (“2019 novel coronavirus disease”[tiab] OR “COVID19”[tiab] OR “COVID-19 pandemic”[tiab] OR “SARS-CoV-2 infection”[tiab] OR “COVID-19 virus disease”[tiab] OR “2019 novel coronavirus infection”[tiab] OR “2019-nCoV infection”[tiab] OR “coronavirus disease 2019”[tiab] OR “coronavirus disease-19”[tiab] OR “2019-nCoV disease”[tiab] OR “COVID-19 virus infection”[tiab]) | 61,770 |
| 3 | (kidney[tiab] OR kidneys[tiab]) | 495,189 |
| 4 | Publication date: 1990/01/01 to 2020/10/30 | - |
| 5 | 1 AND 2 | 1,306 |
| 6 | 1 AND 3 | 106 |
| 7 | 2 AND 3 | 1,179 |
| 8 | 1 AND 2 AND 3 | 34 |
| 9 | 1 AND 2 AND 3 AND 4 | 33 |
Search Syntax of This Study
3.2. Methodological Characteristics
The methodological properties were categorized and summarized in Table 2. Accordingly, 63% of the articles was experimental, 21% review, 5% notes, 5% observational, 3% cohort, and 3% letter to editor. Sampling in 62% of the studies was random, 16% non-random, and 22% was not mentioned.
| Properties of Article | No. (%) |
|---|---|
| Content type of studies | |
| Article | 35 (94) |
| Book chapter | 2 (6) |
| Type of article | |
| Original research | |
| Experimental study | 22 (63) |
| Non-original research | |
| Review | 7 (21) |
| Notes | 2 (5) |
| Observational | 2 (5) |
| Cohort | 1 (3) |
| Letter to editor | 1 (3) |
| Year of publication | |
| 1990 - 2010 | 17 (46) |
| 2010 - 2020 | 20 (54) |
| Population | |
| Human | 10 (24) |
| Animal | 13 (31) |
| Cell | 19 (45) |
| Statistical Society | |
| Coronavirus | 2 (5) |
| Kidney | 1 (3) |
| Coronavirus and kidney | 34 (92) |
| Sample size, number of article | |
| ≤ 50 | 12 (31) |
| 51 - 150 | 6 (16) |
| ≥ 150 | 1 (3) |
| Not mentioned | 19 (50) |
| Age | |
| Human | |
| Under 1 year | 1 (3) |
| Over 1 year | 1 (3) |
| Animal | |
| Under 1 month | 9 (23) |
| Over 1 month | 2 (5) |
| Not mentioned (cell, review, and others) | 25 (66) |
| Type of sampling | |
| Random sampling | 23 (62) |
| Nonrandom sampling | 6 (16) |
| Not mentioned | 8 (22) |
The Properties of Extracted Articles
4. Discussion
4.1. The Role of Coronavirus in the Inducing Kidney Disorders
The link between pathogenic disorder induced by virus and kidney dysfunction is very difficult. The serologic diagnosis, detection of virus-specific antigens, and human antibodies are the main basic problems in establishing glomerular structure destruction. Concentrations of viral antigens in tissues are higher than in the bloodstream, where complex antigens are present with their own antibodies. Different mechanisms differ in the operation of viral nephropathy. In acute glomerulonephritis, viral infection causes the proliferation of glomeruli to release cytokines by altering proliferation. If the virus is quickly cleared, most cases of reversible nephropathy develop. Chronic glomerulonephritis provides steady viral infection with persistent antigenic instigation, thereby producing antibodies and forming immune complexes. Viral proteins cause inflammatory kidney disease by producing some of the intermediates that induce sclerosis and glomerulopathy. Glomerulonephropathy induced by the viral hepatitis B is the most frequent and well known in which immune complexes are formed. Viral rabies infection can make glomerular disorders. There are several laboratory tests for the detection of viral nephropathy, such as molecular tissue analysis. The important mechanisms that induce viral nephropathy are renal virus tropism, immune systems abnormalities, and multi-organ destruction. Acute renal failure can be produced by hantaviruses and coronaviruses, along with acute respiratory syndrome (29, 30). Endemic Balkan nephropathy (EBN) is a renal disease that happened in Bulgaria, Romania, and the former Yugoslavia. These patients are mainly rural people for over 40 years. The main problem in these patients is renal dysfunction without hypertension, which eventually leads to end-stage renal failure. Evidence of viral involvement has been published according to the histological and epidemiological results. It was revealed that EBN was induced by the coronavirus (31). Coronaviruses are pulmonary pathogens in humans that produce one-third of pulmonary tract infections. Also, these viruses can cause diarrhea, and recently, two of them have been isolated from multiple sclerosis. Primary renal tissue culture was performed as implantation of tissue obtained in five EBN patients with urinary tract tumors. Some of the biopsy specimens in incubation with the prolonged culture at 33°C showed a coronavirus virus (EBNV) that was related to human fibroblast cells and cytopathogenic viral cells. Also, in neutralization experiments, EBNV failed to respond to antivirals against these viruses. Using EBNV hyperthermic serum, cytoplasmic immunoreactivity positive for cells infected with human Cv-OC43, HCoV-229E, and Tanjong golden village (TGV) coronavirus were significantly observed in kidney biopsies of EBNV patients. A coronavirus is said to be involved in the etiology of the disease (32).
4.2. COVID-19 and Kidney Injuries
SARS-CoV-2 can enter the kidney cells via interaction with the ACE-2 receptor and priming the cell protease TMPRSS2. This virus has the glycoprotein spike around its membrane, which attaches to the ACE2 receptor binding domain on the membrane of the target cell and diffuse inside the cell. Hence, SARS-CoV-2 induces kidney injuries. COVID-19 increases kidney disorder due to the cytopathic effect of SARS-CoV-2 on kidney epithelial cells and cytokine storm syndrome (CSS). Other renal pathological symptoms of COVID-19 are hypoxia, persistent hypotension, impairment of microcirculation, acute tubular necrosis induced by cytotoxicity or CSS, and kidney proximal tubule injury (33-35).
4.3. Mechanism of Renal Disorders Induced by Coronavirus Except for ACE2 Receptor
Viral bronchitis is a disease of the upper respiratory tract accompanied by urinary tract in chickens. The high mortality is sometimes caused in young chickens due to renal pathology caused by nephropathogenic strains (36). Aminopeptidase N (APN) belongs to the family of membrane-bound metalloproteases. It is expressed in epithelial cells from proximal renal tubules and other tissues, including granulocyte and monocyte tracheal cells, fibroblasts, CNS synaptic membranes, and end-intestinal ciliates and respiratory tracts. Also, the main factor of APN induced pathogenesis is a disturbance in tissues (37). Kidney cells are sensitive to SARS-CoV and support SARS-CoV replication (38). Kidney cell studies have shown that kidney cellular assessment of chicken is useful for the count of viruses, such as coronavirus (39). In the Middle East, health authorities learned that hCoV induces acute respiratory failure and affects the kidneys as an additional role that leads to acute renal failure. The new beta coronavirus was isolated from virus-infected persons in Jeddah, Saudi Arabia, and was called hCoV-EMC (40).
5. Conclusions
The present scoping review showed that coronaviruses such as HCv-OC43, HCoV-229E, and TGV were observed in kidney biopsies of patients, which causes several kidney injuries, including glomerulonephropathy and proximal tubule destruction along with acute respiratory syndrome and tract infection due to inducing renal inflammation and APN expression. Also, it was demonstrated that SARS-CoV-2, as a coronavirus, induces similar pathological effects in the kidney through ACE2 receptor expression and interaction. Therefore, according to the similar effects of SARS-CoV-2 with other coronaviruses in inducing kidney damages, it is suggested that this type of coronavirus may produce its pathological effects via the expression of APN in addition to the ACE2 receptor; however, future experimental and clinical studies are required to clarify this recommendation.