Dear Editor,
Human T-cell leukemia virus type 1 (HTLV-1) is a causative agent of HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) and the adult T-cell leukemia/lymphoma (ATLL) (1). There are 20 million HTLV-1 infected individuals worldwide. Of which, approximately 2% to 4% of HTLV-1 infected cases were developed to ATL; although, 90% of HTLV-1 infected individuals remain as an asymptomatic carrier (ACs) during their lives (2, 3).
ATLL is a progressive lymphoid malignancy, which characterized with an uncontrollable proliferation of CD4+ T cells after a long-lasting period of infection with HTLV-1 in ACs cases (3). The main mechanism of ATL remains unknown; however, it is suggested that cytokines and the immune system play a key role in the development of ATL (3, 4). Due to the existence of several inquiries regarding the ATL pathogenesis; this study was done for the analysis of the main changes in ATL patients compared to ACs via transcriptomic information during a system biology report.
In this study, the differentially expressed genes (DEGs) were retrieved from gene expression omnibus (GEO) datasets (accession number: GSE19080). DEGs were limited to immune-system, apoptosis, cell cycle, and cell growth was analyzed using Benjamini-Hochberg FDR-adjusted P < 0.05 in two different groups of ATLs and ACs individuals. Then, the protein-protein interaction network (PPIN) for significant genes was constructed via STRING online server. Finally, the signaling network for ATLs was proposed for completion of ATL pathogenesis model.
According to our analysis, there are over-expression of different genes including NF-kB, mTOR, PI3K/Akt, transactivation factor, T cell surface molecule as well as anti-apoptotic genes in ATLs; whereas downregulation of IFNG, Caspase, Foxp3, JAK-STAT, or cytokine such as TGF-β in this group. Several of these changes are predictable, for example, downregulation of IFN-γ production followed the destruction of functionality T cells; or decline of TGF-β in ATLs, which inhibits by HTLV-1 (HTLV-1 is inducing production of T cells whereas suppresser effects of TGF-β on T cells proliferation). In addition, our analysis showed that Foxp3 was downregulated in ATLs; given that T regulatory cells are limited T cell proliferation. Therefore, the expression levels of Foxp3 should be down-regulated in ATLs, which was confirmed in this study. Moreover, Janus tyrosine kinases (JAKs) is cause to proliferation or NF-kB and IL-2 was essential for T cell activation and proliferation. Therefore, JAK-STAT, NF-kB signaling pathway, and IL-2 production should be over-expressed in ATLs for T cells proliferation and develop to ATL, which is confirmed in our data analysis (Table 1) (2-5). According to the review of the literatures, Tax is induction of the transcription factors such as CREB, SRF, and AP-1, which is confirmed in our analysis (4, 5).
| Gene Symbol | Description | Function | ATLs | ACs |
|---|---|---|---|---|
| BIRC5 | Baculoviral IAP repeat containing 5 | Apoptosis inhibitor | -0.38 | -0.59 |
| CDC2 | Cell division cycle 2 | Cell proliferation | -0.46 | 0.21 |
| CDKN2A | Cyclin dependent kinase inhibitor 2A | Cell proliferation | 0.26 | -0.13 |
| KCNAB1 | Potassium voltage-gated channel subfamily a member regulatory beta subunit 1 | Potassium channel | 0.31 | 0.16 |
| CREB1 | CAMP responsive element binding protein 1 | Transactivation | -0.54 | 0.001 |
| CD25 | Interleukin 2 receptor subunit alpha | IL-2 receptor | -0.53 | -1.07 |
| NFKBIE | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | Pro- inflammatory response | 0.24 | 0.25 |
| mTOR | Mechanistic target of rapamycin kinase | Cell survive | 1.15 | 0.37 |
| Jak1 | Janus kinase 1 | Inflammation | 0.11 | 0.04 |
| TP53 | Tumor protein P53 | Tumor suppressor | 0.74 | -0.26 |
| PI3K | Phosphatidylinositol-4,5-bisphosphate 3-kinase | Cell proliferation | 0.08 | -0.02 |
| C-myc | C-myc myelocytomatosis viral oncogene | Cell proliferation | -0.78 | -0.72 |
| CCR5 | C-C motif chemokine receptor 5 | Fusion co-receptor | 0.06 | 1.09 |
| CDK6 | Cyclin dependent kinase 6 | Cell cycle regulators | 0.31 | 0.41 |
| Jun | Jun proto-oncogene | Transcription | -0.25 | 0.08 |
| CDKN1A | Cyclin dependent kinase inhibitor 1A | Cell cycle regulators | 0.07 | -0.004 |
| IRF-1 | Interferon regulatory factor 1 | Transactivation | 0.48 | 0.63 |
| SYK | Spleen associated tyrosine kinase | Inflammation | 0.09 | -0.85 |
| NFKB1 | Nuclear factor kappa B subunit 1 | Inflammation | 0.70 | 1.27 |
| TGFB | Transforming growth factor-beta | Suppression of immune-system | -0.06 | 0.23 |
| Foxp3 | Forkhead box P3 | Transcription | -0.49 | -0.07 |
| TRAF | TNF receptor associated factor | Inflammatory response | 0.11 | -0.024 |
| ATF3 | Activating transcription factor 3 | Transcription | 0.07 | 0.3 |
| CREB | cAMP response element binding protein | Transcription | 0.57 | 0.30 |
| CXCR4 | C-X-C motif chemokine receptor 4 | Viral receptor | 3.4 | 2.96 |
| EF2 | Eukaryotic translation elongation factor 2 | Transcription | 0.23 | -0.72 |
| STAT6 | Signal transducer and activator of transcription 6 | Transcription | -1.26 | -0.19 |
| IL-15 | Interleukin 15 | Inflammation | -0.25 | 0.13 |
| SRF | Serum response factor | Transcription | 0.45 | 0.51 |
| BCL2 | B-cell lymphoma 2 | Cell survive | 0.77 | 1.13 |
| CASP9 | Caspase 9 (apoptotic inducer) | -1.27 | -0.98 | |
| IFNG | Interferon gamma | Inflammation | 1.76 | 1.78 |
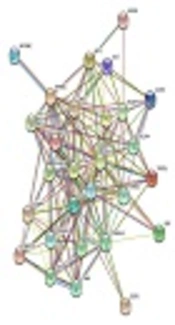
According to PPIN, PI3K/Akt, mTOR, JAK-STAT, NF_kB, and transactivation genes have central roles in ATL pathogenesis, which are located in central nodes. Cytokines, cyclin, T cell surface molecules, and anti-apoptosis genes are located in external nodes, which influenced by central nods (Figure 1).
Based on the signaling network, HBZ and Tax can promote T cells for cell growth and proliferation using several signaling pathways including NF_kB, PI3K, and MAPK signaling pathway, which lead to ATL during lengthy induction by HTLV-1 (Figure 2).
In summary, there is limited information regarding ATL pathogenesis; KEGG pathway (hsa05166) is not enriched enough and future investigation for ATL pathogenesis is needed. This information can be useful for the development of diagnosis, monitoring, and treatment of ATL patients. According to the present analysis, the immune-system changes, particularly cytokines, can have influenced several vital signaling pathways such as NF-kB, MAPK, and PI3K-Akt/mTOR signaling pathways, which regulate cell proliferation and is responsible for developing to adult T-cell leukemia/lymphoma.