1. Background
Myocardial infarction (MI) is an event that occurs in several conditions as a result of several reasons like elevating inflammatory cytokines (1), and percutaneous coronary intervention (PCI) has been suggested to be a good treatment in reducing both short- and long-term death, non-fatal reinfarction, and stroke (2). On the other hand, Clopidogrel is one of the most common antiplatelet drugs used for the prevention of ischemic vascular atherosclerotic disease, acute coronary syndrome (ACS), as well as prevention of Stent thrombosis (3-5). Despite the benefits of using the clopidogrel, some patients experience recurrent ischemic events (6). Platelet reactivity assays showed inter-individual variability in the biological response to clopidogrel (7). Genetics, diabetes mellitus, obesity, smoking, and many drugs are the most important factors which may paly roles in different responses of patients to clopidogrel (8-10). Clopidogrel is a prodrug that is used orally and rapidly absorbed through the stomach. Approximately 85% of the absorbed clopidogrel is hydrolyzed and inactivated by plasma esterases and the reaming is converted to an active metabolite in a two sequential oxidative reaction hepatic cytochrome P450 cytochrome (CYP450) isoenzymes including CYP3A4, CYP3A5, and CYP2C19 (11). Since the enzymatic activity of CYP2C19 depends on the CYP2C19 genotypes, genetic variation within CYP2C19 gene causes variable clopidogrel response. There are more than 25 known variant alleles for encoding CYP2C19 gene (12). The CYPC19*1 allele is the wild type form of the CYP2C19 that encodes the normal functional enzyme, while CYP2C19*2 is the most reported loss-of-function allele in different populations with an allele frequency of 13% - 15% in Caucasians, 18% in African-American and 29-30% in Asian ethnicities (12, 13). Furthermore, CYP2C19*3 is another major loss-of-function allele which influences the pharmacokinetic response to clopidogrel (11). Some studies showed a higher rate of major cardiovascular events after treatment with clopidogrel in the patients carrying these loss-of-function alleles (5, 14, 15). The importance of CYP2C19 genotyping has been established, especially in populations with a high prevalence of *2 and *3 alleles by the United States Food and Drug Administration (FDA) (16, 17). Thus to determining the allele and genotype frequencies of CYP2C19 gene in patients undergoing PCI, genotyping is recommended for application in the prognosis of the response to the treatment and prevention of the complications of stent restenosis (18-21).
2. Objectives
In this study, we determined the frequencies of CYP2C19 polymorphisms in patients who received Drug-Eluting Stents (DES) following PCI who lived in the southwest of Iran.
3. Methods
3.1. Patient Selection
This cross-sectional study was conducted by 102 patients referred to PCI from the department of Cardiology of Imam Khomeini and Golestan University Hospitals in Ahvaz, from Khuzestan province of Iran. Then PCI was performed for all patients with drug-eluting stents. All the patients received aspirin 80 - 325 mg daily for one week before PCI and did not receive thienopyridine derivatives in one week prior to the enrollment.
3.2. Collecting Data
Standardized questionnaires were used to collect data about demographic characteristics, laboratory data, clinical and procedural information of the patients included age, sex, ethnicity, exposure to tobacco smoke, diabetes mellitus, hypertension (systolic blood presser ≥ 140 mmHg or diastolic blood pressure ≥ 90 mmHg), dyslipidemia (LDL-C ≥ 100 mg/dL), body mass index (BMI), white blood cell (WBC) count, platelet (PLT) count, diameter and length of stent, and LVEF < 45%.
3.3. Ethics Committee
All data and sample collection for this study were approved by the Ethics Committee of Ahvaz Jundishapur University of Medical Sciences (REC.ajums.ac.ir 1394.469). Informed consent was taken from each participant and all of them explicitly provided permission for collection of clinical data and genotyping analyses.
3.4. DNA Extract
Genomic DNA was extracted from EDTA-containing whole blood samples using phenol-chloroform method.
3.5. CYP2C19 Genotyping
Here, CYP2C19 genotyping was carried out by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. Two specific PCR reactions for CYP2C9*2 and CYP2C9*3 variant alleles were conducted in parallel for each specimen in a final volume of 10 µl using primers and conditions that previously described by Zendehdel et al. (22). Subsequently, CYP2C9*2 PCR product was digested with SmaI and CYP2C9*3 PCR product with BamHI overnight at 37ºC. The digested products were visualized on a 2% agarose gel stained with ethidium bromide.
3.6. Statistical Analysis
Continuous variables are presented as mean ± SD. Categorical variables are reported as counts (percentage). The gene counting method was used to estimate alleles and genotypes frequencies. CYP2C19 allele and genotype frequencies were analyzed using χ2 test. Differences in allele and genotype frequencies between patients of the present study were measured by Fisher exact test. Data analysis was performed by SPSS (version 22.0) software (SPSS, Inc., Chicago IL, USA). A P-value < 0.05 was considered statistically significant.
4. Results
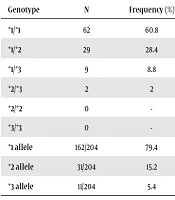
Demographic, clinical, laboratory, and procedural information of the patients are shown in Table 1. Overall, 102 patients (mean age = 65.2 ± 10.8 years) participated in this study. Genotype and allele frequencies of the CYP2C19 polymorphisms are shown in Table 2. The allele frequency of CYP2C19*1, CYP2C19*2, and CYP2C19*3 were 79.4%, 15.2%, and 5.4%, respectively. Also, CYP2C19*1/*1 genotype was recorded in 60.8% of the patients. In addition, 28.4%, 8.8%, and 2.0% of the subjects showed CYP2C19*1/*2, CYP2C19*1/*3, CYP2C19*2/*3 heterozygote genotypes, respectively. The CYP2C19*2/*2 and CYP2C19*3/*3 genotypes were observed in none of the patients.
| Demographic Characteristics | Values |
|---|---|
| Age, y | 65.2 ± 10.8 |
| Sex | |
| Male | 58 (56.9) |
| Female | 44 (43.1) |
| Race | |
| Persian | 69 (67.6) |
| Arabian | 33 (32.4) |
| BMI, kg/m2 | 25.5±3.3 |
| Clinical information | |
| Smoking | 29 (28.4) |
| HTN (BP ≥ 140/90 mmHg) | 27 (26.5) |
| Diabetes mellitus | 47 (46.1) |
| Hyper lipidemia (LDL-C ≥ 100 mg/dL) | 35 (34.3) |
| Laboratory data | |
| WBC (1000/mL) | 6.9 ± 2.2 |
| PLT (1000/mL) | 308 ± 90 |
| Hb, g/dL | 12.4 ± 1.9 |
| Procedural information | |
| LVEF < 45% | 8 (7.8) |
| Length of drug-eluting stent, mm | 20.4 ± 6.1 |
| Diameter of drug-eluting stent, mm | 3.5 ± 1.3 |
Abbreviayions: BMI, body mass index; Hb, hemoglobin; HTN, hypertension; LVEF, left ventricular ejection fraction; PLT, platelets; WBC, white blood cells
aValues are expressed as mean ± SD or No. (%).
| Genotype | N | Frequency (%) | 95% CI |
|---|---|---|---|
| *1/*1 | 62 | 60.8 | 50.6 - 69.0 |
| *1/*2 | 29 | 28.4 | 18.6 - 39.6 |
| *1/*3 | 9 | 8.8 | 3.9 - 15.7 |
| *2/*3 | 2 | 2 | 0.0 - 5.3 |
| *2/*2 | 0 | - | - |
| *3/*3 | 0 | - | - |
| *1 allele | 162/204 | 79.4 | 70.6 - 86.9 |
| *2 allele | 31/204 | 15.2 | 20.2 - 41.2 |
| *3 allele | 11/204 | 5.4 | 5.9 - 17.6 |
Abbreviation: CI, confidence interval for variant allele frequency
5. Discussion
In this study, the frequencies of CYP2C19 polymorphisms were evaluated in Iranian (southwest) patients who underwent PCI and received DES. The results of this study show that the overall relative frequency of CYP2C19*2 alleles is high.
Saber et al. (23) investigated CYP2C19 allele and genotype frequencies on 691 individuals in a multiethnic Iranian population using experimental and computational approaches. The mean frequencies of CYP2C19*2 and CYP2C19*3 alleles were calculated as 0.125 [99.9% CI, 0.112 - 0.139] and 0.006 [99.9% CI, 0.004 - 0.009], respectively by a cumulative meta-analysis performed in the study.
Comparing the studies in different Iranian populations with other populations showed that CYP2C19 allele frequencies in Iranian population are different from Asian ethnicities and are in compliance with African and Caucasian ethnicities (23).
Although based on the results of studies in different populations CYP2C19 genotyping is recommended to use a proper dosage of clopidogrel, some new studies demonstrated contradictions in this regard (17, 18, 20). A recent study by Mahdieh et al. (17) determined CYP2C19, CYP3A5, CYP3A4, and ABCB1 polymorphisms in 388 Iranian patients undergoing PCI treated with clopidogrel during a 6-month period of follow-up. The frequency of CYP2C19*2 was 16.5%. None of the SNPs individually were significantly associated with outcome events. Results of their study showed that combinations of different alleles of genes are involved in pharmacokinetic variability and joint factors are important; so they concluded that genotyping and analyzing an individual variant may not be as straightforward in risk assessment and pharmacogenetics.
Nozari et al. (21) in a case-match study assessed the role of CYP2C19*2 polymorphism in the occurrence of in-stent restenosis during a 1-year follow-up period in Iranian patients who underwent PCI. The results of this study showed no significant association with in-stent restenosis one year after PCI in patients with CYP2C19*1*2 genotype. In another study, Namazi et al. (24) evaluated the impact of P2Y12, CYP3A5, CYP2C19, and environmental factors on the clopidogrel response variability in 112 Iranian patients after PCI. No significant associations between clopidogrel responsiveness and polymorphisms of CYP2C19, CYP3A5, and P2Y12, as well as environmental factors, were shown (P > 0.05).
5.1. Conclusions
The results of this study showed a high prevalence of CYP2C19*2 polymorphism in the population of Khuzestan province located in the southwest of Iran. The frequency of CYP2C19*1/*2 genotype is compatible with the majority of the Iranian population and more similar to Caucasian populations. Since there is controversy in clopidogrel treatment strategy for patients who carry at least one of the non-functional CYP2C19 alleles, further studies with larger sample numbers should be performed to determine the prevalence of non-functional alleles in various populations and to attain an agreement about efficient treatment strategy.