1. Background
One of the most common malignant tumors in women is cervical cancer (CC), which has been diagnosed as fourth cancer in females worldwide (1, 2). Cervical cancer is a component of preventable cancers, and effective screening and appropriate diagnosis and treatment programs have reduced cervical cancer death rates in developed countries (3, 4). Biological, social, and health factors contribute to the formation of this cancer, among which the human papillomavirus (HPV) is known as the most important risk factor for cervical cancer (3-5).
In addition to the HPV virus, genetic factors such as altering the expression of some MicroRNAs (miRNA) also contribute to the development of cervical cancer (6). MicroRNAs can act as oncogenes or tumor suppressors by inhibiting the expression of cancer-related target genes. Besides, functional differences between different types of tumors and various stages of cancer are associated with the expression of microRNAs (7-9).
Moreover, in some studies, the aberrant expression of “mir196a” has been reported in cervical cancer (10). On the other hand, single nucleotide polymorphisms (SNPs) can also be effective in changing the expression of miRNA targets (11). It has also been shown in various studies that miR-196a2 C.T (rs11614913) is involved in various cancers, so we also explored it in this study (12). On the other hand, tumor necrosis factor α (TNF-α) is one of the pro-inflammatory cytokines, which directly contributes to oncogene activation, DNA damage, and, development of cervical lesions (13-15).
2. Objectives
In the present study, the molecular analysis of rs11614913 polymorphism from the miRNA196a gene and its relationship with TNF-α gene expression in cervical cancer was investigated in the Iranian population.
3. Methods
In this study, 52 formalin-fixed paraffin-embedded (FFPE) tissue samples from patients with pre-cancerous and cancerous lesions of cervical tissue and 50 healthy tissue samples were obtained from Khatam Hospital in Tehran. The specimens were examined histopathologically and confirmed by a pathologist.
3.1. DNA Extraction
After deparaffinization of blocks, DNA was extracted by DNA extraction CinnaGen Inc. according to the manufacturer’s protocol and stored at -20ºC.
3.2. Tetra-ARMS PCR Reaction for Proliferation of rs11614913 Polymorphism
Genotype for SNP miR-196a rs11614913 C/T was amplified by T-ARMS PCR in a thermal cycler (Primus 25, Peqlab, Germany) by CinnaGen kits, as well as synthetic primers synthesized by CinnaGen Co (Table 1). The material was poured into PCR-specific tubes, and one sample was considered a negative control that Instead of DNA, 3.5 μL of distilled water was added. The reaction mixture was prepared in a 25 μL volume and after preparation was transferred to the thermocycler apparatus with the program listed in Table 2. Also, the formation of the desired parts in PCR products was investigated by electrophoresis on 2.5% agarose gel.
Sequences of Primers Used for rs11614913 Polymorphism from miRNA196a-2 Gene
| Segment | Temperature | Incubation Time | Number of Cycle |
|---|---|---|---|
| First denaturation | 94ºC | 5 min | x 1 |
| Denaturation | 94ºC | 45 sec | x 35 |
| Annealing | 60ºC | 30 sec | x 35 |
| Extension | 72ºC | 1 min | x 35 |
| Final extension | 72ºC | 10 min | x 1 |
PCR Program for Polymorphism rs11614913 from miRNA196a-2 Gene
3.3. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from the tissues with a CinnaPure RNA kit according to the manufacturer protocols. RNA was quantified by NanoDrop spectrophotometer (BioTek Epoch, America).
Before the reverse transcription reaction was performed, to remove any DNA-contamination, the RNA sample was admixed with a DNA-removing mixture for 2 minutes at 42°C. After DNA removal, the QuantaScript RT enzyme was transcribed, and the RNA was transcribed to the cDNA. Synthesis of cDNA from these genes was performed using the QuantiEect® Reverse Transcription Kit (QIAGEN) and specific stem-loop primers for all types of genes according to the protocol.
3.4. Sequence of Specific Stem-Loop Primers for Genes Includes
SNORD-47 (17): 5’-GTCGTATGCAGAGCAGGGTCCGAGG-TATTCGCACTGCATACGACAACCTC-3’
miRNA196a (18):
5’-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGA-GCCCAACAA-3’
The sequences of primers used are given in Table 3.
Primer Sequences for Real-Time PCR
The expression level of TNF-α and miR-196 in tissues was measured by quantitative Real-time PCR (qRT-PCR) with Snord-47 as the internal reference. Real-time PCR was performed using the ABI 7500 Real-time PCR system (ABI, America) and SYBR GREEN PCR Master Mix (Takara) kit. The amplification conditions for were qRT-PCR were: initial denaturation-95ºc for 5 min, denaturation- 95ºc for 50 sec for 35 cycles, annealing- 60ºc for 40 sec for 35 cycles, extension-75ºc for 1min for 35 cycles and final extension-72ºc for 8 min. The relative expression of miRNA was calculated using Ct values. The difference between the threshold of the desired gene and the internal control gene (housekeeping) can be achieved by obtaining the relative expression of the gene by the 2-ΔΔCt method. Finally, for data analysis, Epi Info software (version 7.1.3.10) and MedCalc (Version 19.2.0) were used.
4. Results
4.1. Tetra-ARMS PCR Results
The frequency of genotypes from rs11614913 polymorphism of the miRNA196a-2 gene was evaluated by using the Tetra-ARMS PCR method. First, the rs11614913 polymorphism of the miRNA196a-2 gene was amplified in all specimens using the PCR method. The desired fragment is 362 bp for miRNA196a-2, which is performed using foreign primers and acts as an internal control in the PCR method, with 362 bp bands present in all samples. According to the statistical analysis of the rs11614913 polymorphism of the miRNA196a-2 gene, the chance of suffering from those with a CC genotype is more than those with TT genotype. We concluded that the CC genotype was a polymorphism of the miRNA196a-2 gene that predisposed cervical cancer (Table 4).
| Genotype/Allele | Total Number of Patients (N = 52), No. (%) | The Total Number of Healthy People (N = 50), No. (%) | P Valuea | OR 95% CI |
|---|---|---|---|---|
| TT | 4 (7.69) | 9 (18) | 0.128 | |
| CT | 18 (34.61) | 23 (46) | 0.242 | 1.609 |
| CC | 30 (57.69) | 18 (36) | 0.029 | 0.412 |
| CC | 30 (57.69) | 18 (36) | ||
| TT+CT | 22 (42.3) | 33 (64) | 0.002 | 0.414 |
| CC+CT | 48 (92.3) | 41 (82) | ||
| TT | 4 (7.69) | 9 (18) | 0.02 | 0.347 |
| T | 26 (25) | 41 (41) | ||
| C | 78 (75) | 59 (59) | 0.016 | 2.085 |
Frequency of Types of CC, TC, and TT Genotypes from rs11614913 Polymorphism in Healthy and Diseased Patients
4.2. Comparison of TNF-α Gene Expression and miRNA196a Expression
In this method, the normalized CT value is measured relative to an untreated sample, and we also need internal standards, their CT values should be deducted from the CT specimen value (normalization).
The relative difference of the test sample versus the control is calculated by 2-∆∆CT formula:
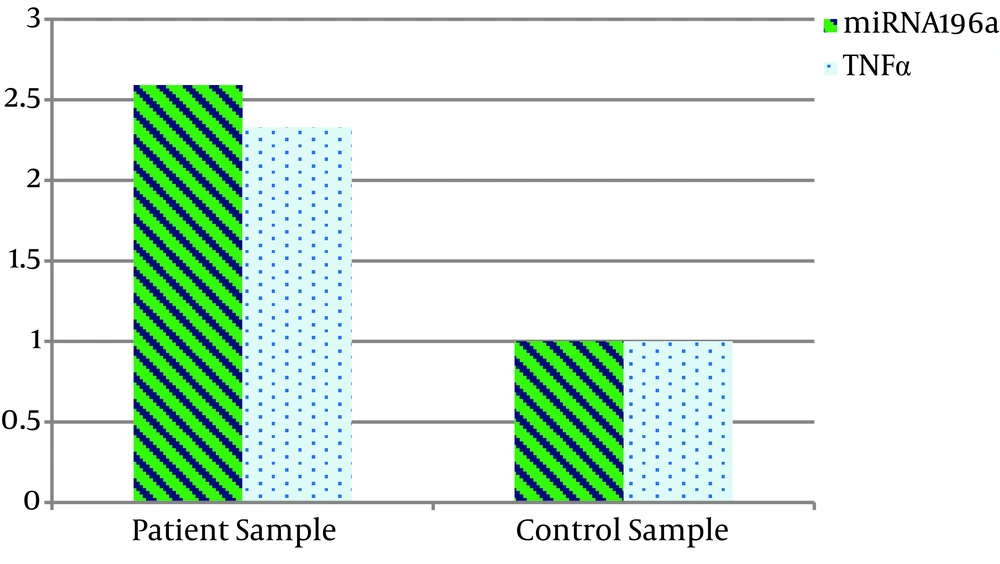
The results of the comparison of miRNA196a and TNF-α gene expression in both healthy and patient groups showed that miRNA196a gene expression was about 2.59 fold, and TNF-α was about 2.33 fold higher in the patients than in the controls (Figure 1).
4.3. The Relationship Between Types of miRNA196a Gene Polymorphisms by TNF-α Gene Expression
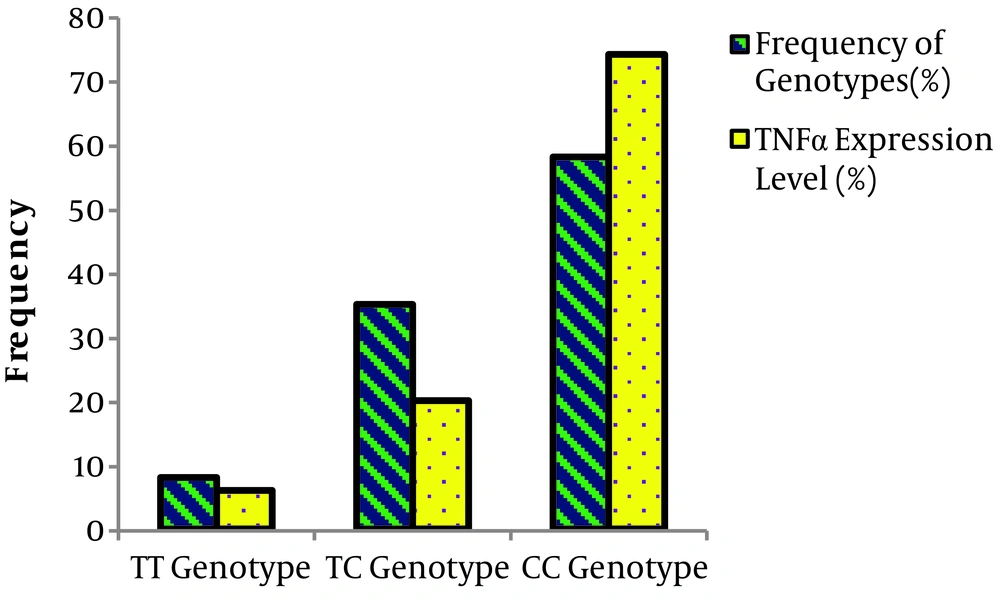
The results showed that there is a significant relationship between the frequency of TT, TC, and CC genotypes and the level of TNF-α expression in the affected individuals (Table 5 and Figure 2). In other words, the CC genotype increases the expression of the TNF-α gene in comparison to the TT genotype of miRNA196a gene polymorphism.
| Genotype/Allele | Frequency of Genotypes (%) | TNF-α Gene Expression (%) | OR 95% CI | P Valuea |
|---|---|---|---|---|
| TT | 8 | 6 | 0.282 | 0.287 |
| TC | 35 | 20 | 0.136 | 0.003 |
| CC | 58 | 74 | 9.35 | 0.0006 |
| T | 25 | 11 | ||
| C | 75 | 89 | 0.371 | 0.01 |
Relationship Between miRNA196a Polymorphism Genotypes and the Expression of the TNF-α Gene
5. Discussion
This study involves several research aspects. First, the molecular analysis of rs11614913 polymorphism from miRNA196a gene in cervical cancer, and second, the study of the expression of miRNA196a and TNF-α genes in cervical cancer, and finally, the relationship between TNF-α gene expression and genotype polymorphisms rs11614913 from miRNA196a gene in cervical cancer.
Numerous studies have shown the regulatory role of miRNAs in various types of cancer, in which the role of miRNA196a in various types of malignancies has been proven, and rs11614913 as a polymorphism of miRNA196a gene in some cancers is effective (22, 23). The role of TNF-α in various types of cancer has also been shown in numerous studies (13-15). Therefore, a multifaceted study of these factors can help identify biomarkers that are effective in cervical cancer.
Our results in this study showed that the C allele was associated with an increased risk of cervical cancer, and, on the other hand, a significant correlation was found between CC genotype and rs11614913 polymorphism from miRNA196a gene in the cervical cancer sample. Previous studies have also demonstrated the relationship between this SNP and other cancers. For example, one study demonstrated that carriers of the variant homozygote CC of miR-196a-2 rs11614913 were more likely to develop epithelial ovarian cancer (EOC) compared with wild‑type homozygote TT and heterozygote CT carriers (23).
Bodal et al. (24) showed that heterozygous genotype of miR196a2 and combined polymorphism of miR-146 and miR196a2 genes were associated with increased risk of breast cancer in North Indian Women. Guan et al. (7) found that statistically significant associations with HPV16-positive SCCOP and survival for Hsa-mir-196a2 rs11614913. Also, for the first time Tian et al. found that variant genotype CC of miR-196a2 rs11614913 was associated with a significantly increased risk of lung cancer in the Chinese population (25).
However, some studies, unlike our study, did not find a significant relationship between this polymorphism and cancer. For example, Hashemi et al. found no statistically significant association between miR-196a2 rs11614913 variant and PCa risk in the Iranian population (26). Very limited studies have been conducted on the rs11614913 polymorphism of the mir196a gene in cervical cancer. In 2017, the results of Srivastava et al. in India showed that rs3746444 T/C gene polymorphism of the Hsa-miRNA499 gene had a significant relationship with the risk of cervical cancer, while polymorphisms of miRNA146a and miRNA196a2 did not show any association with cervical cancer. Although in the same study of Srivastava, miRNA196a2 gene polymorphism in smokers has a high risk of cervical cancer, no significant changes in the risk of cervical cancer are observed for other polymorphisms (11).
Genetic polymorphism involves a person’s susceptibility to cancer and heavily dependent on environmental mutagens. The reason for this difference in research findings can be in different types of races and geographical conditions, as well as differences in the type of life in each region (27). On the other hand, in the present study, the increase in the expression of the miRNA196a gene in people with cervical cancer has been observed in comparison with healthy subjects, and various studies have also shown similar results to this study. In 2016, a study by Yang et al. In China showed increased a196miRNA gene expression in cancerous ovarian tissue and benign ovarian tissue compared with normal ovarian epithelial tissue (28).
Another study by Liu et al. in China revealed that the serum level of miRNA196a was elevated in patients with cervical cancer compared to healthy subjects. Moreover, the serum level of miRNA196a was associated with CIN grade and various clinical parameters important for cervical cancer (29). Hou et al. also observed a significant increase in miRNA196a in people with cervical cancer compared to healthy subjects (30). Therefore, the expression of the mir196a gene can be considered an effective biomarker for cervical cancer.
In the present study, like mir196a, the expression of TNF-α has also increased in cervical cancer, and other studies have also shown the importance of TNF-α in cancers. For example, a study by Govan et al. in South Africa found that TNF-α plays an important role in all phases of cervical cancer (31). Also, in a study by Al Obeed et al., an increase in TNF-α expression in cancerous tissues was observed in patients with colorectal cancer compared with healthy subjects (15). Furthermore, in another study conducted by Piura et al. in Israel, there was an increase in the expression of TNF-α in cancerous tissues of ovarian cancer compared with healthy tissues (32).
5.1. Conclusions
Considering the results of this study, the expression of TNF-α gene in patients was higher than the control group, and further studies showed that CC genotype increases the expression of TNF-α gene by miRNA196a gene polymorphism than TT genotype. It can be concluded that TNF-α can be a good biomarker for cervical cancer, and more studies with larger sample sizes and different ethnicities can help confirm this acclaim.