1. Background
Cancer is the third leading cause of death in Iran after cardiovascular disease and accidents and is also the second leading cause of death worldwide (1, 2). Acute lymphocytic leukemia (ALL) that is known as acute lymphoblastic leukemia is the most common type of leukemia, which is more common among people under the age of 15 years in Iran (3). In Iran, as a developing country, cancer is on the rise. Leukemia is about 8% of all human cancers (4). Leukemia, based on the rate of progression of disease to acute and chronic and also based on the involved cells to lymphoid and myeloid, is divided into two types (5). ALL is a cancer of the lymphoid line of blood cells. It is characterized by the development of large numbers of immature lymphocytes. The accumulation of these cells in the lymphatic tissues causes these organs to enlarge. Also, a significant increase in lymphocytes leads to a decrease in the number of other blood cells such as red blood cells and platelets, and this imbalance in blood cells leads to anemia, bleeding, and non-coagulation. This type of leukemia progresses rapidly and is typically fatal within weeks or months if left untreated (6).
Various studies have shown that the time between the appearance of the ALL disease and its spreading is very short, and the most common type of leukemia is in children, which is often seen in children between the ages of 2 and 6. ALL can also occur in adults, though the chance of a cure is greatly reduced, and people over the age of 75 are more vulnerable to the disease (7).
Some genes or proteins as tumor markers are somehow involved in the progression of cancers. Molecular markers can play an important role in determining the nature of leukemia-related tumors and can be used as a diagnostic adjuvant along with precise pathological methods. These markers must be simple and fast in terms of technique and interpretation of results and have high sensitivity (8).
Apolipoprotein A1 (ApoA1) is a protein and the major component of HDL particles that, in humans, is encoded by the ApoA1 gene. This gene is located on the 11th chromosome with its specific location, 11q23-q24. It contains four exons. Some studies elucidated that genetic changes in the ApoA1 gene led to triglycerides increasing and HDL decreasing (9-12). Numerous studies have shown that lower HDL and higher triglycerides are associated with cancers (13-17). Naik demonstrated that serum cholesterol, HDL, and LDL are significantly decreased and associated inversely with the incidence of cancer, whereas triglycerides were significantly elevated in cancer patients compared to the normal population (13), which is in agreement with the findings of Musolino et al. (14) and Fiorenza and colleagues (15). The relationship between hypocholesterolemia and the degree of maturation of leukemic blast cells in acute myeloid leukemia was demonstrated in the other studies (16). Also, Peterson et al. demonstrated that hypocholesterolemia in cancer patients may be caused by elevated LDL receptor activates in malignant cells. The inverse association between cancer and serum cholesterol may reflect a physiological response to early undiagnosed stages of cancer (17).
Two siblings with HDL deficiency and no plasma apolipoprotein A-I (Apo A-I) were found to be homozygous for a cytosine deletion in exon 3 of Apo A-I gene (c.85 del C, Q5FsX11). This mutation causes a frameshift leading to a premature stop codon and abolishes the synthesis of ApoA1. In two other unrelated subjects, HDL deficiency was due to heterozygosis for a nucleotide substitution in exon 4 of Apo A-I gene (c.494 T > G, L141R) (18). A family with hereditary non-neuropathic amyloidosis has reported that hereditary hepatic and systemic amyloidosis caused by a new deletion/insertion mutation in the ApoA1 gene in which 12 residues have been deleted and two residues inserted in the amyloidogenic NH2-terminal fragment. The 5’ end of exon four of the ApoA1 gene reported as abnormal, thereby producing a PCR product of 361bp fragment as the deletion and 396bp as the insertion mutation (19).
2. Objectives
In the present study, the role of different modes of ApoA1 gene polymorphisms in patients with ALL, and the frequency of different alleles of insertion/deletion mutation of the ApoA1 gene was investigated.
3. Methods
The present study was a case-control (retrospective) study consisting of a case study and a control population. The 50 blood samples from patients with ALL and healthy subjects were taken. The patients were diagnosed by a physician specialized in diseases, and they were under treatment (chemotherapy of maintenance phase in Ali Asghar Hospital affiliated to Iran University of Medical Sciences and Imam Khomeini Hospital in Tehran). Among the healthy people who referred to the health centers in the west of Tehran affiliated to Iran University of Medical Sciences, the healthy subjects were selected through simple randomization. The study population was examined from March 2017 to January 2017, and written informed consent was obtained from all patients before the sampling. The required sample size was obtained to study 25 patients with ALL and 25 healthy subjects based on the sampling size of similar studies and limited access to patient samples (9, 20). The alpha level was set at 0.05, and the study strength was 80%.
DNA was extracted from blood samples by salting out method. A rapid, safe, and inexpensive method was developed to simplify the deproteinization procedure. This method involves salting out of the cellular proteins by dehydration and precipitation with a saturated NaCl solution according to the following procedure: A total of blood samples were collected and kept at 4°C. Then 1.5 mL of cell lyses buffer was added to 500 μL of each blood sample. The cell lysates were digested overnight at 37°C with 0.2 mL of 10% SDS and 0.5 mL of a protease K solution. After digestion was complete, 1 mL of saturated NaCl (approximately 6M) was added to each tube and shaken vigorously for 15 seconds, followed by centrifugation at 12,000 rpm for 15 minutes. The precipitated protein pellet was left at the bottom of the tube, and the supernatant containing the DNA was transferred to another tube. Exactly, 2 volumes of absolute ethanol were added, and the tubes were inverted several times until DNA precipitated. The precipitated DNA strands were removed and transferred to a 1.5 mL microcentrifuge tube containing 100 - 200 pL TE buffer. The DNA was allowed to dissolve for 2 hours at 37°C before quantitating. Qualitative analysis of the DNA extracted was determined with the help of Agarose gel 1.5% of electrophoresis, and its concentration was measured using Nanodrop (21, 22).
Genotype distribution and allele’s frequencies were determined by the Gap-PCR method. Using a pair of primers presented in Table 1, which was designed and optimized with the help of Gene Runner software and the BLAST method of the National Website of the Biotechnology Information Center (http://www.ncbi.nlm.nih.gov). The Gap-PCR method is a technique used to detect large mutations. The oligonucleotide primers used in this technique are designed to cover both sides of the removed area. If deleted, the two primers come close together, and DNA replicates. This method is considered a simple, fast, and inexpensive alternative compared to Southern blotting and fluorescent in situ hybridization (FISH) methods (23, 24).
| Sequence (5’ → 3’) | Length | Tm | GC | Name | |
|---|---|---|---|---|---|
| Forward primer | AGGACCTGCTGGGGACTAAAG | 21 | 61.1 | 57.1 | ApoA1 |
| Reverse primer | TGAGAAACCTGCTGCCTCTGC | 21 | 62.9 | 57.1 |
The promoter area to the intron 2 of the 856 bp ApoA1 gene were amplified through Gap-PCR under the following conditions: 100 nanograms of genomic DNA were added to the reactive mixture containing Taq buffer (10 mM tris-chloride, 50 mM KCL, 0.1 triton X100, 25 μL) (2x Master mix Red, Taq DNA Pol) and then 5 pM/μL was added from each of the primers and the final volume was increased to 50 μL (20). PCR was performed in 35 cycles with optimal conditions according to Table 2.
| Number of Cycles/Steps | Steps of Gap-PCR | Temperature (ºC) | Time |
|---|---|---|---|
| 1/1 | Primary denaturation | 95 | 5 min |
| 35/2 | Denaturation | 95 | 0.5 min |
| Annealing | 62 | 0.5 min | |
| Extension | 72 | 1.5 min | |
| 1/3 | Final extension | 72 | 5 min |
Finally, the Gap-PCR product was detected based on electrophoresis on agarose gel and ethidium bromide staining against ultraviolet light.
Moreover, SPSS software version 16 was used to record data and statistical analysis. A chi-square test was used to check the frequency of the genotype and its differences in the healthy subjects and patients. The confidence level in all experiments was 95%, and the level of significance was less than 0.05.
4. Results
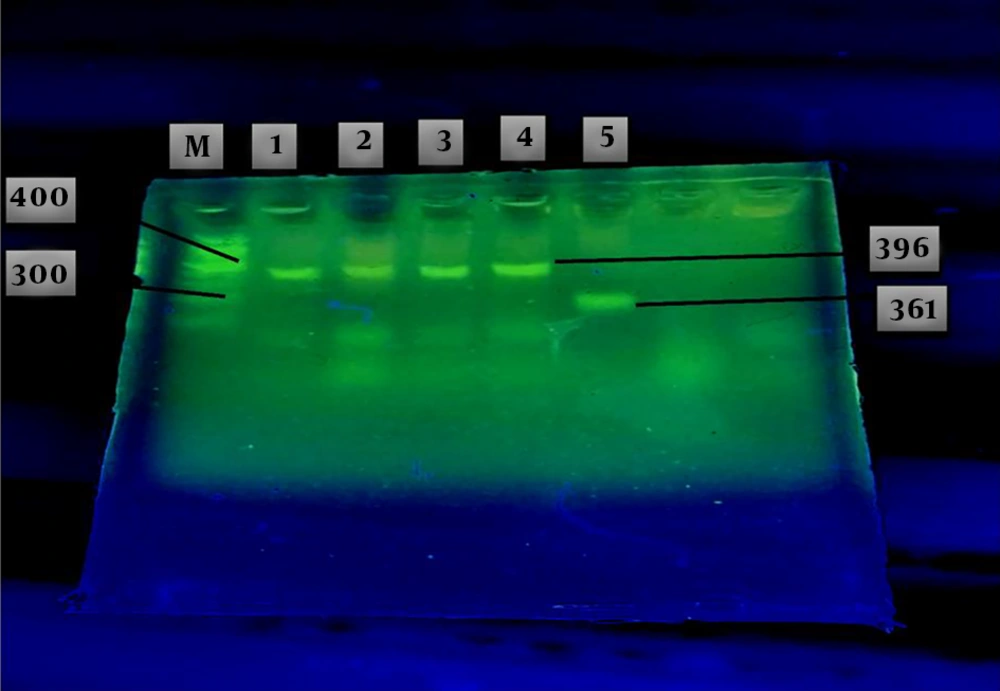
The results of agarose gel electrophoresis of the GAP-PCR products related to the insertion and deletion mutations of the ApoA1 gene in patients are shown in Figure 1. Two fragments were achieved, one of which was 361 bp related to the deletion mutation of the ApoA1 gene, called allele D, as a genotype of DD, and the other fragment, with 396 bp, was related to the insertion mutation of the ApoA1 gene, named I allele, as a genotype of II (Figure 1). Heterozygote state was not observed in these cases like other subjects.
Agarose gel electrophoresis of the GAP-PCR products related to Insertion and Deletion mutation of the ApoA1 gene in the patients with ALL (lanes 1 - 5). A fragment with 396bp yielded in lanes 1 - 4 related to the insertion mutation as a genotype of II. The other fragment with 361 bp (lane 5), related to deletion mutation as a genotype of DD. Markers of known length were run in lane M.
The results of the genotype distribution among the patients showed that there were 21 homozygotes with genotype II and 4 homozygotes with genotype DD and nobody with heterozygote genotype DI. Among the healthy subjects, 22 homozygotes with genotype II and 3 homozygotes with genotype DD, also nobody with heterozygote genotype DI was found. Based on the relevant statistical analysis, despite the high frequencies of both genotypes in males than in females, there was no significant difference between genotype distribution of healthy subjects and patients (P = 0.516) (Table 3).
| Sex/Group | II Genotype | II Percent | DD Genotype | DD Percent | Total | Percent | P |
|---|---|---|---|---|---|---|---|
| Male | 0.516 | ||||||
| Patients | 17 | 34 | 3 | 6 | 20 | 40 | |
| Healthy | 15 | 30 | 3 | 6 | 18 | 36 | |
| Total | 32 | 64 | 6 | 12 | 38 | 76 | |
| Female | |||||||
| Patients | 4 | 8 | 1 | 2 | 5 | 10 | |
| Healthy | 7 | 14 | 0 | 0 | 7 | 14 | |
| Total | 11 | 22 | 1 | 2 | 12 | 24 | |
| Total of male and female | 43 | 86 | 7 | 14 | 50 | 100 |
The results of sex-based genotype distribution showed that ALL disease had a higher relative frequency percentage in males than females. However, there were no significant differences between healthy subjects and patients (P = 0.508) (Table 4).
| Gender | Cumulative Frequency | Relative Frequency Percentage | Relative Frequency | Absolute Frequency | P |
|---|---|---|---|---|---|
| Male | 0.508 | ||||
| Patient | 20 | 40 | 0.4 | 20 | |
| Healthy | 38 | 36 | 0.36 | 18 | |
| Female | |||||
| Patient | 43 | 10 | 0.1 | 5 | |
| Healthy | 50 | 14 | 0.14 | 7 | |
| Total | 50 |
The results of frequency subjects of the sample population based on age are presented in Table 5. The percentage of absolute frequency of 6 - 8 years was the lowest, and 8 - 10 years was the highest rate compared to the other subjects. The results of genotype frequency and its relationship with ALL based on age showed that there was no significant difference between healthy subjects and patients (P = 0.662) (Table 5).
| Age (Year) | Samples Mode | Absolute Frequency | Genotype II | Percent II | Genotype DD | Percent DD | P |
|---|---|---|---|---|---|---|---|
| 6-8 | 7 | Patient | 3 | 6 | 0 | 0 | 0.662 |
| Healthy | 4 | 8 | 0 | 0 | |||
| 8-10 | 15 | Patient | 4 | 8 | 1 | 2 | |
| Healthy | 9 | 18 | 1 | 2 | |||
| 10-12 | 14 | Patient | 7 | 14 | 1 | 2 | |
| Healthy | 4 | 8 | 2 | 4 | |||
| 12-14 | 14 | Patient | 7 | 14 | 2 | 4 | |
| Healthy | 5 | 10 | 0 | 0 | |||
| Total | 25 | Patient | 21 | 42 | 4 | 8 | |
| 25 | Healthy | 22 | 44 | 3 | 6 |
5. Discussion
Studies on different modes of a genetic predisposition suggest that factors such as genetic polymorphisms may indicate individual differences in susceptibility, premature onset and progression of cancer, and even drug resistance. Acute lymphocytic leukemia (ALL) is more than forty cases per million children per year (5). Eighty percent of ALL cases occur in children and 20 percent in adults, and the disease is more common in males than in females that is in agreement with the findings of this study (4). The disease does not have a good prognosis in adults; despite significant advances in treatment, its mortality is more common in adults. In various studies, the rate of mortality has been stated to be 25% to 40% (7). According to research conducted in Iran, this type of leukemia accounts for the largest percentage of all cancers in children under 15 years of age (25). Previous studies have reported that ApoA1 is a potential marker for many types of cancer, including breast and pancreatic cancer (26, 27). Another study reported an association between HDL-C/ApoA1 plasma levels and the risk of developing a wide range of cancers (28). Sheikhha et al. (29) reported no significant relationship between the polymorphism of the ApoA1 gene and the risk of diabetes that is consistent with the results of the present study.
Owing to the fact that in a few studies conducted in this regard (26-28), no significant differences were observed, and therefore, the results of this study are consistent with the results of similar studies.
While many studies have indicated that polymorphism of ApoA1 does not have a significant relationship with ALL disease in different communities, the opposite is also true in one study (30) that this observed inconsistency is due to differences in the related allele frequency connected to the genetic background of different populations.
Owing to the fact that the risk of ALL disease is influenced by a combination of genetic and environmental factors, for this reason, different results have been obtained among the populations in some studies.
However, in order to check the serum level of ApoA1 protein, a specific kit was needed, and the preparation of this kit was not possible. Therefore, it was not possible for us to measure serum levels of it. Nevertheless, owing to the non-significant difference of this gene in the healthy subjects and patients, it does not seem necessary to measure the relevant protein.
The results of this study and other similar studies (26-28) showed that ApoA1 polymorphism does not play a role in ALL incidence in the community. Although the ApoA1 gene plays an important role in fat metabolism and lymphocyte metabolism, it does not play a significant role in the processes involved in the reproduction, hematopoiesis, and natural immune system (leukemia). One important reason for this could be the small sample size of the study population. Finally, it is suggested that in order to resolve the existing ambiguities, in future studies, sequencing should be performed in all areas of the ApoA1 gene, and the interventions of ApoA1 gene polymorphisms in a larger Iranian population should be investigated.
In this study, there was no significant difference between the healthy subjects and patients in terms of the frequency of alleles and genotypes, which was consistent with some studies (27, 29). One of the reasons that can be stated is that the genetic association is population-dependent (31). In this regard, the subjects studied are important reasons. Thus the subjects were randomly selected in this study, while matching has been done in many other studies so that individuals are matched in terms of various factors such as Body Mass Index (BMI), fat index, gender, and age characteristics.
5.1. Conclusion
Based on the result of this study, it can be concluded that ApoA1 gene polymorphisms were not associated with ALL and were not effective in the formation or exacerbation of it in this population. Also, it could not be identified as a prognostic marker.
It should be noted that these findings are the first report of the degree of association of ApoA1gene insertion/deletion polymorphisms with ALL, and it is recommended that this study should be performed on more individuals.