1. What Are Exosomes?
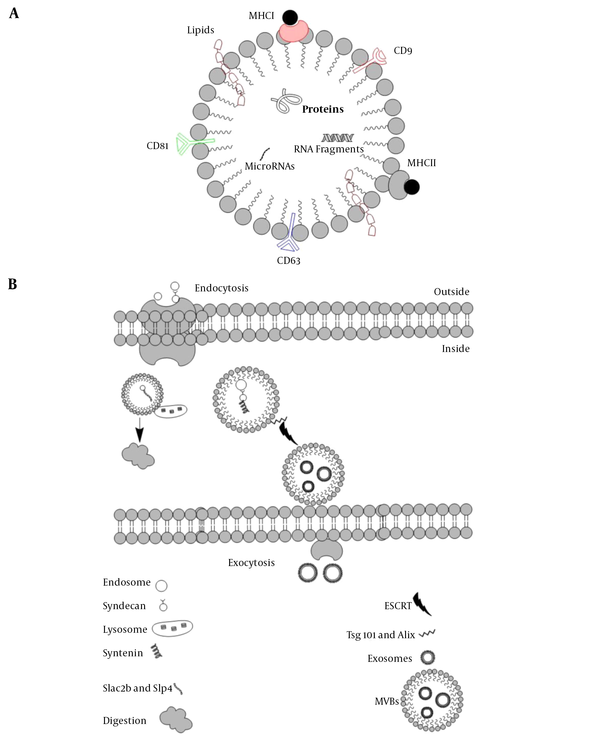
About five decades ago, exosomes were first characterized in the extracellular milieu. The word “exosomes” was first used in 1970 when two scientists were working on reticulocytes. They found uniform sacs released by the cells (so-called exosomes), in sharp contrast with the material that internalizes inside the cells via endocytosis, However, exosomes were known to be the waste materials released by cells. The term “exosomes” first appeared in a scientific paper in the 1980s. For many years they were considering as waste disposal until the last decade when they became the center of interest, due to their role in cell communication as well as their role as the carrier for small sequences of DNA and RNA, lipids, and even proteins (e.g., MHC I, MHC II, CD9, CD63, CD81) (Figure 1) (1).
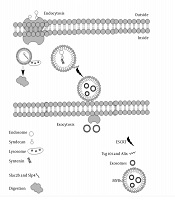
The basic structure of exosomes A, EVs can carry macromolecules such as lipids, proteins, and RNAs. In addition to unique CD markers, it can express HLA on its surface; B, The fate of endosomes is schematically represented, EVs decorated with Slac2-Slp4 and attached to the lysosome and digested or marked with syndecan-syntenin and later attached to ESCRT to release as exosomes.
Exosomes are known as vesicles with plasma membrane that trace back to the conjugation of numbers of multivesicular bodies (MVBs). Lysosomes then digest MVBs or are released in the extracellular membrane in the form of intraluminal vesicles (ILVs). These ILVs liberated from the cells are known as exosomes. Endosomal sorting complexes required for transport (ESCRT) and its subunits, ESCRTI, ESCRTII, and ESCRTIII help the conformational changes in the cell membrane for exosomes release (2). There are some other accessory proteins in the ESCRT complex, such as Tsg101 and Alix. A comparison of exosomes and MVBs based on their characteristics, size, morphology, protein marker, origin, mechanism of discharge, and compositions is provided in Table 1. Based on the original classification of the extracellular vesicles (EVs), they are known as apoptotic bodies (50-5000nm), microvesicles (100-1000nm), and exosomes (40 - 100 nm).
| Exosomes | Microvesicles | References | |
|---|---|---|---|
| Initiation | ILVs release from the cell membrane | Plasma membrane bulging | (2) |
| Mechanism of release | ESCRTs pathways | Ca2+ pathway | (3) |
| Membrane contains | Cholesterol, phosphatidylserine, lipid rafts | Cholesterol, phosphatidylserine, lipid rafts, diacylglycerol | (1) |
| Carrying | Proteins, mRNA, miRNAs | Proteins, mRNA, miRNAs | (3) |
| Size, nm | 40 - 100 | 100 - 1000 | (4) |
Comparing Exosomes and Microvesicles Based on Size, Origin, Mechanism Release and Action, and Macromolecules Contents
Five Rab GTPases contribute to exosome formation in HeLa cells. Rab27a and Rab27b are of crucial importance in exosome secretion (5). It seems that endosomes containing syndecan heparan sulfate bind to syntenin, and this complex later attaches to ESCRT and its accessory protein Alix. In this process, c-Terminal fragments of 6 (ARF6) along with phospholipase D2 (PLD2) are the leading players in transforming the endosomal syndecan-syntenin complex into MVBs that eventually are released as exosomes (6).
2. Routine vs. Improved Exosomes Detection
The most common methods to isolate and characterize the exosomes are ultracentrifugation (UC) and western blotting (WB). In the case of the UC, such methods can be implemented based on various criteria, including density (7) and coprecipitation (8). Microfluidics is a newly applied method to detect exosomes. It has the same disadvantage as the two last gold standard methods (9). A three-dimensional pattern of nanoparticle microfluidics chips detects the exosomes in plasma of ovarian patients with higher precision. The nanoparticle weaved in broken twill weave, or herringbone pattern increases the surface area for detecting a small amount of sample, 2 µL of plasma. Biomarkers of the vesicles are the foundation of some other techniques to isolate the exosomes. These methods are based on the affinity of the macromolecules on the surface of the exosomes. For such cases, titanium oxide is one of the most common techniques that binds to the phospholipid membrane of the exosomes and other extracellular vesicles of patients’ serum (10). Implementing the monoclonal antibody technique can improve the exosomes’ isolation a monoclonal cells. Monocolonal antibodies attached to the microbeads are useful in detecting several biomarkers of cancer patients in their exosomes. Currently, the exosomes with the purity of 93% isolated from glioblastoma cancer patients can be used as a biomarker of cancer (11). Using these isolation methods requires a large number of samples and implementing a tedious procedure. Also, the efficiency and sensitivity of these two methods are very low (9). Sized exclusion chromatography (SEC) is another method to isolate the exosomes and other extracellular vesicles based on the size. Columns of the chromatography are filled with pores gels that capture the exosomes based on their size. SEC demonstrates higher sensitivity in comparison to the UC, but quantitatively capture lower numbers of exosomes (12). Different microscopical techniques are introduced to analyze the EVs, especially exosomes, based on the shape.
Scanning electronic microscopy (SEM), transmission electronic microscopy (TEM) (7), and Atomic force microscopy (AFM) (13) have been employed in various exosomes studies. For the quantitative detection of exosomes, two techniques are introduced, which are based on laser light scattering and browning motions. The first method is dynamic light scattering (DLS), and the second is the nanoparticle tracking analyzer (NTA). DSL only analyzes the distribution of the exosomes, but NTA examines both concentration and the size of the exosomes (14). Another technique that uses voltage is tunable resistive pulse sensing (TRPS) that measures the size and concentration of the exosome (15). To test the quality of the macromolecules like proteins in exosomes, techniques such as WB, enzyme-linked immunosorbent assay (ELISA) (16), and flow cytometry are the most prevalent methods. In the case of the RNA content of the exosomes, ethanol precipitation technique is widely used. The RNA is suspended aquatic milieu and isolated in columns filled with silica gels. Later, polymerase chain reaction (PCR) amplifies the RNA sequences and sends it for sequencing (17).
3. MicroRNA Fragments of Exosomes
EVs also contain microRNAs. A recent study on EVs of mesenchymal stem cells revealed that microRNAs affect angiogenesis in cardiac muscles, in particular, prevent apoptosis in cardiomyocytes and reduce collagen production by cardiac fibroblasts. Moreover, these microRNAs can upregulate angiogenesis in human umbilical vein endothelial cells (HUVECs). Thus, EVs microRNA has an impact on different cell signaling pathways, including Wnt, transforming growth factor-beta (TGF-β), and platelet-derived growth factor (PDGF) intracellular cascades. Plasma-derived exosomal microRNAs have been used as a diagnostic tool for type 1 diabetes mellitus (T1DM). At least seven different microRNAs are altered in T1DM patients. Blood samples taken from lung cancer patients and healthy donors showed that microRNA-181b-5p and microRNA-21-5p are highly expressed in the first group. EVs MicroRNAs-possess neurodegenerative effects that notoriously impact the progression of Alzheimer’s disease (AD) by affecting Tau and amyloid precursor proteins (18). Moreover, EVs microRNAs induce inflammation via activation of toll-like receptors (TLRs) activation. Thus, blood circulating exosomes that can pass through the blood-brain barrier (BBB) can be an excellent diagnostic and prognostic tool in neurodegenerative diseases, such as multiple sclerosis (MS). The sequencing of EVs microRNAs has indicated the presence of entirely different microRNAs in relapsing versus progressing MS patients (19). MicroRNAs 21, 133a, and 181b are highly expressed in EVs in the serum of colon cancer patients. Interestingly, the expression of microRNA 21 in serum EVs is an early sign of the progression of colon cancer (20). Moreover, the diagnostic power of EVs microRNA is proved in chronic kidney disease (CKD), related to microRNA-16 expression in the urine of these patients (21).
4. Exosome-Based Vaccines
Unfortunately, the intravenous delivery of EVs for therapeutic purposes has not yielded outstanding results due to their precipitation in the kidney, liver, and spleen. However, EVs engineering opens new therapeutic avenues. Recently, the assembly of EVs carrying glycosylphosphatidylinositol (GPI)-anchored nanobodies was successful for anchoring protein antibodies and other substances for targeted-exosome therapy. These nanobodies do not modify the physical characteristics of EVs and can specifically target tumor cells if fused with ligands. Bioengineering of exosomes by transfecting cells with membrane proteins, such as G proteins and HLA-A, had potential effects on producing EVs loaded with these proteins. Dendritic-derived exosomes (Dex) and tumor-derived exosomes (Tex) have been used for therapeutic purposes since two decades ago. In addition to major histocompatibility complex molecules (MHCs), Dex bears adhesion molecules to find their targets. Also, these dendritic cells derived exosomes have co-stimulatory molecules and cytosolic factors for exosomes production in targeted cells (22). Moreover, exosomes extracted from M1 pro-inflammatory macrophages were injected to stimulate the lymph node residential DCs, cytotoxic T cells, and macrophages. Furthermore, EVs improve the activity of lipid calcium phosphate (LCP) nanoparticles encapsulated Tyrosinase-related protein-2 (Trp2) vaccine during melanoma cell growth (23).
5. Exosomes and Drug Delivery
Liposomes molecules that are very similar to exosomes, which are artificially made to deliver drugs. These structures have a bilayer phospholipid with hydrophobic and hydrophilic parts. The hydrophilic drugs loaded into the liposome at the hydrophilic part while the hydrophobic drugs load into the hydrophobic one. Another essential feature of the liposome is that it can be engineered to carry proteins.
Due to its lipidic structure, the liposomes can pass the cell membrane and induce essential effects to the host cells. Despite all the advantages of liposomes, toxicity, and removal by the immune system are its main disadvantages. Liposomes are coated with PEG to shield them from the immune system attack and opsonization. However, exosomes can better deliver drugs because they do not trigger an immune response and are not toxic. Naturally, they can carry RNA and other biological informative material, proteins, and lipids, so they can easily be engineered. Furthermore, based on their origin, exosomes can target different tissues. For example, DCs genetically manipulated to release exosomes containing proteins, including rabies virus glycoprotein (RVG) and lysosome-associated membrane protein 2B (LAMP2B), can pass from the BBB. Electroporation is used to load the exosomes with exogenous small interfering RNA (siRNA). After passing from the BBB, SiRNA down-regulates the targeted RNA inside the microglia, oligodendrocytes, and neurons (24). It also showed that food-derived exosomes, such as grape exosome-like nanoparticles (GELNs), target intestinal stem cells and protect against dextran sulfate sodium-induced colitis, thus increasing the response of the intestine to pathological disorders (25). Drugs can be loaded into exosomes either passively or actively, depending on the amount that should be delivered. Passive loading can be done simply by co-culturing drugs with exosomes directly, or drugs can be incubated with donor cells (e.g., mesenchymal stem cells), and then exosomes can be purified from the medium that cells were incubated. In the case of dynamic loading, the exosome and the drug of interest must undergo different treatments and procedures. Exosomes naturally carry microRNAs and siRNAs. The exosomal envelope protects the RNAs against rupture and RNases. As exosomes pass the cell membrane and even the BBB, they can be excellent candidates for microRNA and siRNA delivery. In recent years, CD34 stem cells have been used to produce exogenous Cy3 dye-labeled pre-microRNA precursors that can pass through other cell membranes and regulate gene expression. Exosomes can also be loaded with epidermal growth factor receptor (EGFR) microRNA and target breast cancer in Rag2-/- mice (26). In addition to all the advantages of exosomes for delivery of small interfering RNA or other nucleic acids, still, some technical problems should be addressed. A useful source of cells that produces a high number of copies of the viable exosome still represents a big hurdle. Additionally, exosome, microRNA production, and purification are a tedious procedure as Lamp2b is the main protein to express on exosomes, a new way to manipulate other proteins for targeted delivery crucial. Despite the effectiveness of the delivery using exosomes, they are continually removed by the kidney and the liver. Lack of a competent way to deliver proteins via exosomes is a crucial challenge. Recently, it has been shown that vesicular stomatitis virus glycoprotein (VSVG) conjugated with a fluorescent reporter can be used to load the wanted protein into exosomes of HEK293 cells and increase the delivery of the proteins of choice. Fluorescent microscopy was used to track exosomes efficiently (27).
6. Exosomes and Disease Treatments
All around the world, many people are infected by bacterial, fungal, and viral infections. Viruses use the exosomes wisely to spread their territory to the new cells. Viral infections send their proteins and genetic information through exosomes to infect more cells without rupturing the host cells and going into lysogenic phases. Transactivating RNA (TAR-RNA) packed into exosomes of the human immunodeficiency virus (HIV) can raise the amount of intracellular level of inflammatory cytokines (28). Parasites also use the exosomes to further get extra benefits from the host. Plasmodium uses a defensive strategy against the host immune system by applying exosomes to attenuate host immunity. Plasmodium-infected cells release exosomes to increase CD40 on the DCs and macrophages. Consequently, through increased inflammatory factors around the area, the parasite will survive. In the case of cancer tissues, exosomes play a critical role in cancer cell proliferation and metastasis (29). Exosomal miRNA released by cancer cells increase the tumorigenicity by the notorious impact on the Dicer protein. After passing from the cell membrane, the exosomes release the miRNAs, and as a result, the new sign of cancer emerges in the cell received the exosomes. Non-small lung cancer cells release exosomes that contain proteins such as EGFR, GRB2, and SRC, which can affect the recipient cells and ignite the metastasis (30). So, targeting the exosomes can be one of the leading areas in the case of treating various diseases such as cancer and viral infections. This targeting can be at exosomes formation level, release, uptake, and trafficking.
The best tactic to prevent exosomes formation is to use natural or synthetic material and using small non-coding RNAs, such as siRNAs or shRNAs. A drug called amiloride that decreases the blood pressure has a critical role in preventing the formation of the exosomes. Amiloride inhibits the ceramide genesis and consequently, blockage of exosomes formation. Amiloride implementation on cancer-bearing mice and human downregulate the metastasis rate by disrupting the development of the exosomes containing associated heat shock protein. Using RNAi GW4869 to prevent ceramide development also showed declining signs of the circulatory miRNAs and inflammatory cytokines in cancer patients (31). As mentioned above, the ESCRT and Rab27 GTPase are critical for exosomes release, so, targeting these proteins using RNAi can be very effective to hamper proliferation and metastasis of cancer cells. One of the most important proteins in up taking the exosomes inside the host cells is Phosphatidylserine. Therefore blockage of this protein is pivotal for diminishing the effects of the disease-causing by exosomes (32). Besides, targeting proteins such as nucleoplasm-specific nuclear exosome targeting (NEXT), that has a key role in exosomal miRNA construction and stability and, as result exosomal trafficking, can decline the effects of the exosomal causing diseases (33).
7. Exosomes in Clinical Trials
Based on what was mentioned before, exosomes are a delivery system for drugs, miRNAs, and (even) DNAs. The most common cells that are implemented to produce exosomes are DCs and mesenchyme stem cells (MSCs). Immune-modulatory effects of MSCs are well known (34), so, these stem cells are a good source of exosomes for attenuating the inflammatory responses in the patients, especially cancer patients. Also, other cells, such as stem cells derived from adipose tissues and HEK293 cells line, are the other two best options that can produce a large number of exosomes (35). In this scenario, all the processes must get along with good manufacturing practices (GMP). For GMP, three main criteria should be considered, first of all, the environment that the cells thrive is critically important for producing high-quality exosomes. The second step to produce high-quality exosomes is the purification of the exosomes. In addition to the traditional method, such centrifugation, which is a laborious method, tangential flow filtration (TFF) is used to purify the exosomes on a large scale and in a shorter time. After purification, the exosomes can be used for clinical trials. DCs derived exosomes that contain the tumor antigen are good candidates for stimulating immune cells against tumor tissues. Exosomes containing MHCII molecules, 8.5 × 1011 to 4.0 × 1013, that derived from DCs practiced on the melanoma and non-small lung cancer cells. Data analyzed in these cases indicated the safety of the procedure in both cases, while patients with non-small lung cancer revealed a large number of T-cells against cancer tissues. Besides, employing the exosomes as a delivery system both for drug and RNAi has been used recently. Applying plant-derived exosomes has some advantages and disadvantages in comparison to animal-derived ones. Exosomes derived from plants don’t have contaminations that can be found in animal sources (36).
8. Biomarkers Based Exosomes and Diagnostics Shortly
However, it is a demanding task to find the difference between exosomes and other EVs. However, as the exosomes are representative of what is going on in the cells, they are valid candidates to be used for therapeutic and diagnostic purposes. It is revealed that breast cancer cells shed a lower rate of the exosomes in comparison to the normal cell line (37). Even the exosomes size defines their fate. And these criteria can be applied for detecting the distention or biomarkers of special tissues. Based on their size, exosomes are divided into three groups, ExoA: less than 75 nm, ExoB: 75 - 100 nm, and ExoC: greater than 100 nm (38). in addition to the size variations, exosomes’ contents also have been used as biomarkers. Exosomes contain CD63 called Exo1, CD9 Exo2, and CD81 Exo3. It is demonstrated that the protein contents of the exosomes demonstrate the originality of them. For example, the exosomes that trace back to the DCs are different than MSCs, based on the protein and nucleic acid contents. The exosomes originated from the brain are known as ExoI, pancreas derived exosomes are called ExoII, and Liver exosomes are ExoIII. The functionality of the exosomes also is divergent. Some of the exosomes induce survival signals on the target cells, Exoα, some induce apoptotic cascade in the recipient cells Exoβ, Exoβ, and some impose immunomodulation effects Exoγ (39).
9. Conclusions
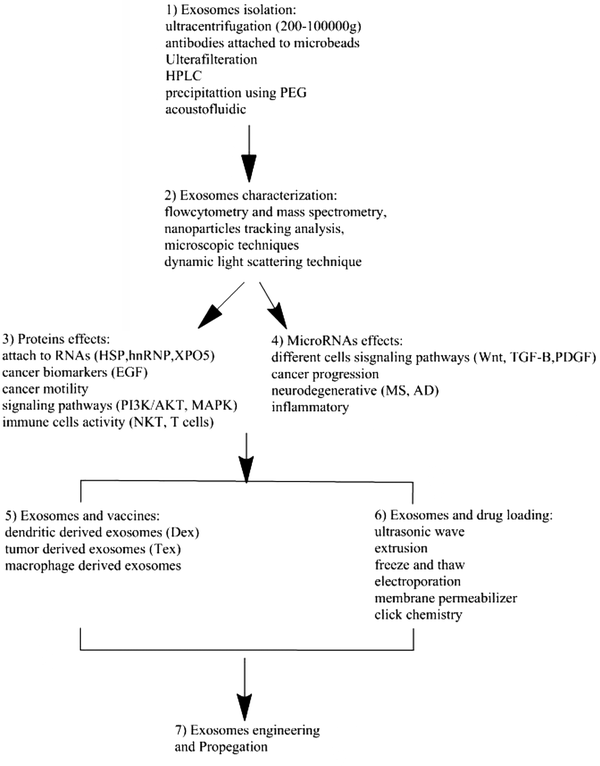
EVs are nanosize particles that are naturally produced by a wide variety of cell lines and tissues. Their ability to cross cell membranes and communicate with other cells makes them ideal candidates to be manipulated for clinical purposes. These nanosize particles that can carry proteins, small RNAs, and lipids are emerging as a new way to treat diseases, including cancer (Figure 2). Therefore, EVs can be perfect carriers of targeted drug delivery.
The diagram shows the use of EVs in the clinic. 1, Isolation of EVs. Methods such as ultracentrifugation; 2, characterization of the physical and chemical properties (FACS and electronic microscopy); 3 - 4, Importance of content of protein and microRNA in EVs for therapy; 5, Use of EVs for vaccination; 6, Methodology such as freeze and thaw and ultrasonic waves are used to load the exosomes with drugs or microRNAs; 7, Importance of EVs for therapeutic purposes.
These proteins express in exosomes; some are structural proteins, and others have functional properties. These Proteins are sometimes cancer biomarkers. By analyzing proteins in the exosomes, it is possible to diagnose cancer. Exosomes released from healthy cells and immune cells have immunogenic roles in the body. In addition to the protein fragments, microRNA packed inside the exosomes are functionally important. Apart from their role in angiogenesis, they can be used as an excellent diagnostic tool to diagnose diabetes, cancer, renal dysfunction, and Alzheimer patients.
Lipid bilayer structure and nanosize character of the exosomes make them the right candidate for drug and nucleic acid delivery. Naturally produced exosomes or exosomes manipulated by assembling proteins on them have therapeutic purposes. Exosomes released by monocytes can be purified and used as a vaccine to stimulate the immune system. In contrast with liposomes, exosomes can circulate in the body without sending a danger signal to the immune system. Furthermore, because of their small size, they can pass BBB easily.