1. Background
Cancer is the second leading cause of death globally behind cardiovascular disease (1). The word cancer is the translation of a Greek word, karkinos, meaning crab, initially used by Hippocrates (370 - 460 BC) (1). With 10.9 million new cases and claiming 6.7 million lives annually, cancer is a dangerous disease for which developing prevention and treatment strategies to manage the disease depends on our understanding of factors that contribute to its development (2). Breast cancer is one of the most common cancers, responsible for approximately 22% of all cancers. Besides, it is one of the leading causes of cancer death in women worldwide (14% of all cancer-related demises) (3). Factors such as age, gender, genetic background, and nutrition are involved in cancer development (4). According to the evidence, 15% - 20% of all cancer cases are due to viruses (5). Viruses are obligate intracellular parasites that can control host cell signaling pathways by producing proteins, which causes the virus to take control of proliferation, differentiation, and apoptosis and escape recognition by the immune system (5). EBV has been demonstrated to be involved in Burkitt’s lymphoma, HPV in cervical carcinoma, and HBV in hepatocellular carcinoma. HHV-8 is a virus that can grow in different cells. Besides, it can induce Kaposi’s sarcoma (6). This virus is a dsDNA virus from the Herpesvirales order, Herpesviridae family, Gammaherpesvirinae subfamily, Rhadinovirus genus.
There are studies that have investigated the association between HHV-8 and breast cancer. Some reported that HHV-8 infection is correlated with breast cancer, while some rejected such association (7, 8). Besides, according to the literature, HHV-8 can immortalize and transform breast cells by inhibiting apoptosis, inducing cell proliferation, enhancing cell survival, increasing angiogenesis, and modulating the immune system. Moreover, the virus can induce tumor genesis by producing interleukin homologs (8, 9). Therefore, fighting against viruses is of particular importance in the body. One of the important functions of the immune system is to fight against viral infections and eradicate cancer cells.
TLRs are proteins that play a vital role in the innate immune system, a function of which is to identify various viral components (10). Currently, 13 types of TLRs have been identified in mammals, of which TLRs 1 - 10 are expressed in humans (11). The human TLR4 gene is located in the cytogenetic locus of 9q33.1 and produces a protein with an alternative name as CD284 (12). TLR4 is positioned in the cytoplasmic membrane and is expressed in both groups of hematopoietic and non-hematopoietic cells (13). One of its functions is to identify the protein structures of viruses, in other words, the protein has an antiviral role (14, 15). Studies have shown that some TLR4 polymorphisms are associated with viral infections (14, 16, 17). According to the evidence, increased expression and activity of TLR4 protein in chronic infectious and inflammatory conditions enhances the progression of cancer (18). On the other hand, some TLR4 polymorphisms have been found to be associated with cancer (19, 20).
2. Objectives
The aim of this study was to examine the correlation between rs4986791 polymorphism in TLR4 gene and HHV-8 infection and their relationship with breast cancer.
3. Methods
3.1. Sample Collection
The present study is approved by the Ethics Committee of the Arak University of Medical Sciences (code: IR.ARAKMU.REC.1395.288). Subjects were sampled among those who referred Ayatollah Khansari Hospital in Arak from November 2016 to April 2017. In total, 80 women with breast cancer and 80 healthy women were recruited. After completing the questionnaire, blood samples (2 mL) of all participants were collected and stored at -20°C for subsequent tests.
3.2. DNA Extraction
DNA was extracted from the whole blood using the Irazola kit (RNA company, Iran) according to the protocol, and the extracted DNA was stored at -20°C until use.
3.3. HHV-8 Detection
The presence of HHV-8 was assessed by PCR using primers used in previous studies (Table 1). The PCR process was performed in a volume of 25 µL containing 10 pmol of each primer (1 µL), 25 µmol of dNTPs (0.4 µL), DNA template 5 µL (50 ng), 10XTaq buffer 2.5 µL (CinnaGen, Iran), 2 mM of MgCl2 (1 µL) and 2 units of Smar Taq DNA polymerase (CinnaGen, Iran), distilled water (15.9 µL). The PCR product was finally electrophoresed on 2% agarose followed by analyzing the results.
| Gene/Virus | SNP | Sequence (5’ to 3’) | Annealing (Co) | Amplicon Size, bp | Reference |
|---|---|---|---|---|---|
| HHV-8 | F: AGCCGAAAGGATTCCACCAT | 59 | 233 | (21) | |
| R: TCCGTGTTGTCTACGTCCAG | |||||
| TLR4 | rs4986791 | External: F:AGTTGATCTACCAAGCCTTGAGT | 53 | 510 | (22) |
| External: R:GGAAACGTATCCAATGAAAAGA | |||||
| Internal: F:GGTTGCTGTTCTCAAAGTGATTTTGGGAGAA | 55 | 407 | |||
| Internal: R:ACCTGAAGACTGGAGAGTGAGTTAAATGCT |
Primer Sequences, Annealing Temperatures and Amplicon Size for HHV-8 Detection and Genotyping of TLR4
3.4. Genotyping of TLR4
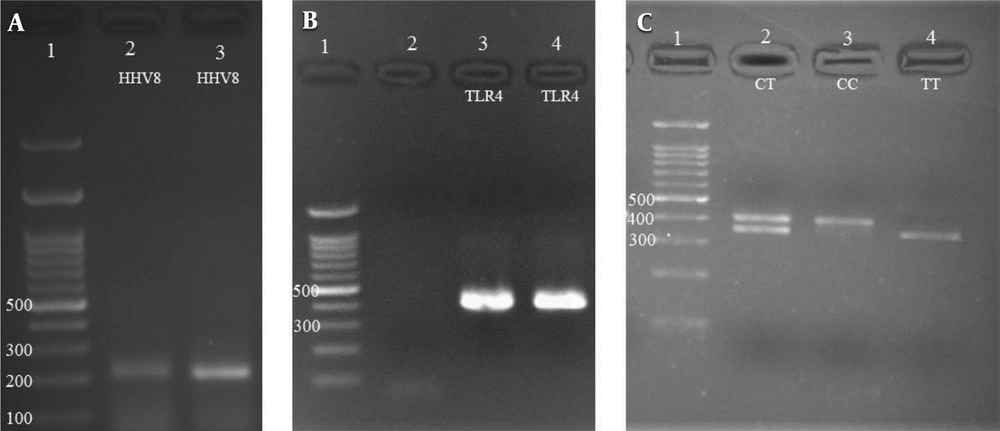
The rs4986791 polymorphism was investigated by nested PCR-RFLP assay using previously-used primers in the literature (Table 1). Primer Blast (www.ncbi.nlm.nih.gov/tools/primer-blast) and restriction enzyme cleavage domain information from the NEB cuter database (http://nc2.neb.com/NEBcutter2/) were used to ensure the performance accuracy of primers. The PCR process was performed in a volume of 25 µL containing 10 pmol of each primer (0.3 µL), 25 µmol of dNTPs (0.4 µL), 2 µL DNA template (50 ng), 2.5 µL of 10XTaq buffer (CinnaGen, Iran), 2 mM of MgCl2 (0.5 µL) and 2 units of Smar Taq DNA polymerase (CinnaGen, Iran) distilled water (18.8 µL). Then, 5 µL of the PCR product was mixed with 17 µL of water, 2.5 µL of the buffer, and 0.5 µL of Hinf I enzyme and incubated at 37°C for 1 hour. Finally, the results of enzymatic digestion were electrophoresed on 3% agarose.
3.5. Physicochemical Analysis of Protein Structure
The physicochemical information of SNP rs4986791 in the mutated and wild-type forms of the TLR4 amino acid sequence were investigated in the Expasy database (https://web.expasy.org/protparam/).
3.6. Statistical Analysis
Data were analyzed by SPSS version 20 using the chi-square test. Statistical significance was considered when P-value < 0.05. The relative odds ratio (OR) was calculated by logistic regression, with a 95% confidence interval.
4. Results
The mean age of patients was 49 years, with the youngest and the oldest subjects aged 26 and 86 years, respectively. Nine patients were younger than 40 years of age and 71 aged over 40 years. Seventy-seven participants were married, and the rest were single or widows. The results of the t-test showed a significant relationship between age and cancer (P < 0.05).
4.1. HHV-8 Detection
Of 80 patients and 80 healthy subjects, 17 (21.3%) and 15 (18.8%) subjects were carriers of HHV-8, respectively (Figure 1A). However, the presence of this virus was higher in cancer patients than in healthy subjects. Statistical analysis showed no significant relationship between HHV-8 infection and breast cancer (P = 0.693). Mean ages of 47 and 46 years were recorded for the patients with HHV-8 infection in both healthy and patient groups and virus-free subjects, respectively. There was no significant relationship between age and HHV-8 infection (P = 0.578).
Of 17 patients with HHV-8, 16 had Grade II and III, of which 8 (27.6%) were with Grade II, 8 (25%) with Grade III, and one (25%) with Grade IV. No significant relationship was found between the presence of virus and cancer grade (P = 0.618).
4.2. Genotyping of TLR4
The rs498679 polymorphism was evaluated in patients and healthy controls (Table 2, Figure 1B, and C). The results of the chi-square test revealed a significant relationship between this polymorphism and breast cancer (P = 0.002). According to the results, the frequency of the T allele was higher in the healthy group, which indicates the association of this allele with breast cancer (P < 0.05). In other words, the T allele increases health phenotype and reduces the risk of cancer (P = 0.000).
Distribution of Alleles and Genotypes Among Patients and Controls
On the other hand, an OR less than 1.0 (0.401) was calculated for the T allele versus that for C. This value was lower and higher than 1.0 in patients and healthy subjects, respectively, indicating a significant relationship, hence the T allele reduces the incidence of a phenotype. The results showed that the T allele reduced the risk of cancer phenotype and increased the incidence of a healthy phenotype and that this polymorphism in the CC raised the chances of cancer development (P = 0.000).
There was a significant association between genotypes and disease. The results indicated that the TC genotype (OR = 1.468, CI = 95% 1.086 - 1.985) could increase the healthy phenotype. On the other hand, the C allele may be associated with cancer due to the higher frequency of CC genotype in patients, because of the increased frequency of CC genotype in patients.
4.3. The Association Between HHV-8 Infection and rs4986791 Polymorphism
Of 15 healthy individuals diagnosed with HHV-8, six and nine subjects had CC (CC is homozygous of allele C) and TC genotypes (CT is heterozygous of alleles T and C), respectively. Of 17 patients carrying the virus, nine and eight subjects had the CC and TC genotypes, respectively. HHV-8 was detected in none of the TT genotypes. According to the results of the statistical analysis, those with the TT genotype (TT is homozygous of allele T) were significantly less infected with HHV-8 than the other genotypes. In other words, individuals with the TT genotype were more immune to HHV-8 infection.
4.4. Physicochemical Analysis of Protein Structure
The physicochemical structure of the protein was investigated two times: once with wild-type allele and natural amino acid, and once with the mutant amino acid sequence for rs4986791 polymorphism. In Table 3, C is the wild-type allele, and T is the mutant of the TLR4 gene polymorphism. No changes were found in the isoelectric pH and instability index in both the mutant and wild-type modes, but the molecular weight, GRAV, and aliphatic index were elevated in the mutant (T) compared to the wild-type mode (C).
Computation of Various Physical and Chemical Parameters in Wiled Type and Mutant Type in TLR4 Protein
5. Discussion
Cancer is the second leading cause of death globally behind cardiovascular disease (1). Breast cancer is one of the most common female cancers (accounts for about one-third of all female cancers) behind lung cancer. Moreover, it’s the most common cause of death in women (23). Several factors contribute to cancer development, including viruses. HHV-8 is one of the pathogenic viruses containing a number of genes homologous to human genes, including cell proliferation, anti-apoptosis, and angiogenesis (24). The tumorigenic potential of this virus is known, particularly in immunocompromised individuals (5). Few studies have investigated the association of this virus with breast cancer. The immune system plays a key role in preventing viral infections. In addition to their role in defense against pathogen invasions, TLRs also play a key role in cancer development through their tumorigenic and antitumor effects (25). The stimulatory effect of TLR in cancer has been proposed as a double-edged sword because it inhibits tumor progression in some cancers and facilitates tumorigenesis and metastasis in others (25). Therefore, studying these biological molecules is important.
Although extensive studies have investigated the association between the Herpesviridae family members and cancer, as noted previously, evidence regarding the relationship between this virus and breast cancer is not sufficient. El-Shinawi et al. (26) examined the presence of HHV-8 in the breast tissue of 135 women with breast cancer (44 women with inflammatory breast cancer (IBC) and 91 individuals with non-IBC). They reported that the HHV-8 virus was detected in only one (out of 91) non-IBC individuals and four (out of 44) IBC cases. In tissues that their viral DNA was confirmed, a higher level of Ki-67 protein expression, a marker of proliferation in proliferating cells, was observed (26). The results indicated a possible functional relationship between the presence of viral DNA and the disease pathogenesis. In other words, this finding suggested the role of HHV-8 in malignancy.
Tsai et al. (27) examined the presence of HHV-8 in 62 women with breast cancer and 60 healthy controls, including 12 patients without cancer, 16 with thyroid cancer, and 32 with fibroid adenomas. Of 62 patients with breast cancer, 28 (43.8%) and 28 (87.5%) of control fibroadenoma subjects carried HHV-8 virus. For patients with thyroid cancer and those without cancer, authors reported no presence of viruses. Comparing those with breast cancer and their healthy controls revealed a significant association between HHV-8 and breast cancer. The comparative comparison of the malignant breast cancer group with those of the benign fibroadenoma group indicated that HHV-8 was more associated with malignant tumors of the breast tissue (27). Both of these studies demonstrated the role of HHV-8 in increasing malignancies, which contradicts our observations. The type of investigated tissue samples may be an important reason for the observed difference. Also, it worth noting that, in the present study, all samples were obtained from patients with stage two cancer or upwards. Mohamed et al. also confirmed the results of El-Shinawi et al. (26) and Tsai et al (9). Another important reason for the observed difference may be the low prevalence of HHV-8 in Iran, as several studies reported a low prevalence of the virus in HIV-infected people in Iran (28-30).
Changes in a nucleotide have different effects on protein structure. Analysis of the physicochemical information of the protein revealed that the isoelectric pH of both genes did not change after the amino acid modification. The GRAV index in the TLR4 gene polymorphism was decreased in the mutant state being somewhat more positive, suggesting a greater tendency for the protein to a nonpolar environment after modification. Also, the aliphatic index was increased in these proteins in the mutant state, which in turn raised hydrophobicity and temperature tolerance of this protein after genome changes and mutations. These findings indicate a change in the protein, which is likely to influence the function of proteins and their affinity to target molecules.
The rs4986791 (C/T) polymorphism frequencies of 80.6% and 62.5% were estimated for the C allele in the patient and control groups, respectively. Based on the statistical calculations, there was a significant relationship between the genotypes and the disease development. Few studies have investigated the association between rs4986791 (C/T) polymorphism and breast cancer; In this line, there are few comprehensive studies on the association between this polymorphism and the development of other cancers.
Kurt et al. (31) demonstrated that individuals with CC genotype were 3.857 times more likely to develop lung cancer compared to those with CT genotype in rs4986791 polymorphism, which is in line with our findings. Another meta-analysis by Khademalhosseini et al. (32), on the role of TLR4 polymorphism in breast cancer, revealed that rs4986791 polymorphism plays a role in breast cancer. They also mentioned the dual role of this polymorphism in carcinogenesis or cancer treatment. The type of ligands can be considered as an important factor in determining the outcome of TLR4 function. Saturated fatty acids, for example, are ligands that may be associated with the negative role of TLR4 in breast cancer. If accompanied by H2O2, TLR4 ligands can stimulate TGF-β1 signaling, which is a critical pathway for the detection of metastases. Overall, TLR4 can be a friend or an enemy of breast cancer. TLR4 can fight breast cancer tumor cells by recognizing breast cancer-related DAMPs and then through the bodily immune responses. On the other hand, up-regulation of TLR4 and overexpression of breast cancer-related DAMPs can alter TLR4 functions resulting in tumor progression (32).
A meta-analysis on rs4986791 by Zhang et al. (33) reported that TLR4 polymorphism is involved in cancer development. Most of these articles (2000 - 2012) concerned cervical and gastrointestinal cancers. A meta-analysis by Ding et al. (19), on 27 articles with a total of 4,416 cancer patients and 7,379 controls established that rs4986791 polymorphisms were associated with cancer, particularly in the Asian population. Both of the above studies are consistent with our findings.
Theodoropoulos et al. (34) examined rs4986791 polymorphism in 261 breast cancer patients and 480 healthy individuals and reported no significant relationship between breast cancer and rs4986791 polymorphisms. Kina et al. (35) examined rs4986791 polymorphism in glioblastoma cancer patients and mentioned no associations between TLR4 polymorphisms and glioblastoma cancer. Differences observed between various studies can be attributed to factors such as study population, sample size, heterogeneity of studied tumors, and the dual role of TLR in the health or carcinogenicity. Inconsistencies in the expression patterns of TLR4 in various cancers can preclude any conclusion about its decisive role in cancer.
5.1. Conclusions
This study demonstrated a significant relationship between rs4986791 polymorphism and breast cancer. Also, it was found that the T allele can increase healthy phenotype and decreases the risk of cancer development. Also, none of the individuals with the TT genotype were infected with the HHV-8 virus, indicating the successful function of the TT genotype in immune responses. Although in the present study we found no significant association between HHV-8 infection and breast cancer, the authors recommend, as found in other investigations, performing further research on HHV-8 infection and increased malignancies in breast cancer, as well as the role of TT genotype in TLR4 rs4986791 polymorphisms in the prevention of infection with different viruses.